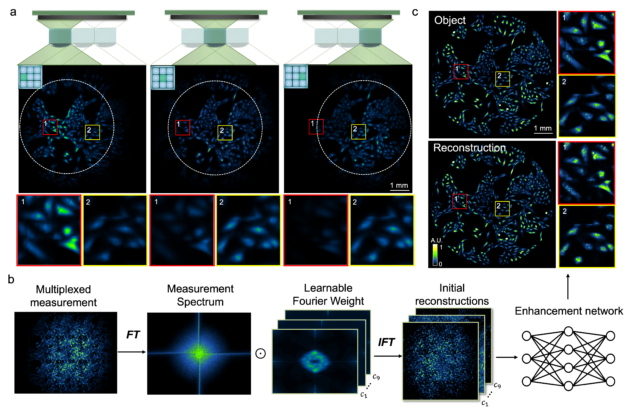
Deep learning for biomedical imaging
Wide-field, high-resolution reconstruction in computational multi-aperture miniscope using a Fourier neural network
Qianwan Yang, Ruipeng Guo, Guorong Hu, Yujia Xue, Yunzhe Li, and Lei Tian
Optica Vol. 11, Issue 6, pp. 860-871 (2024).
⭑ Github Project
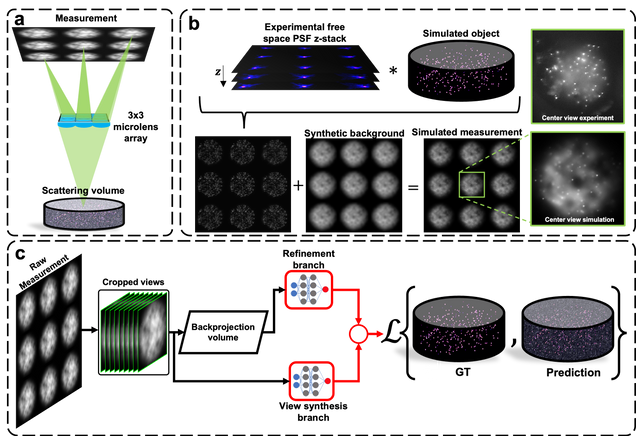
Robust single-shot 3D fluorescence imaging in scattering media with a simulator-trained neural network
J. Alido, J. Greene, Y. Xue, G. Hu, Y. Li, K. Monk, B. DeBenedicts, I. Davison, L. Tian
Optics Express Vol. 32, Issue 4, pp. 6241-6257 (2024).
⭑ Github Project
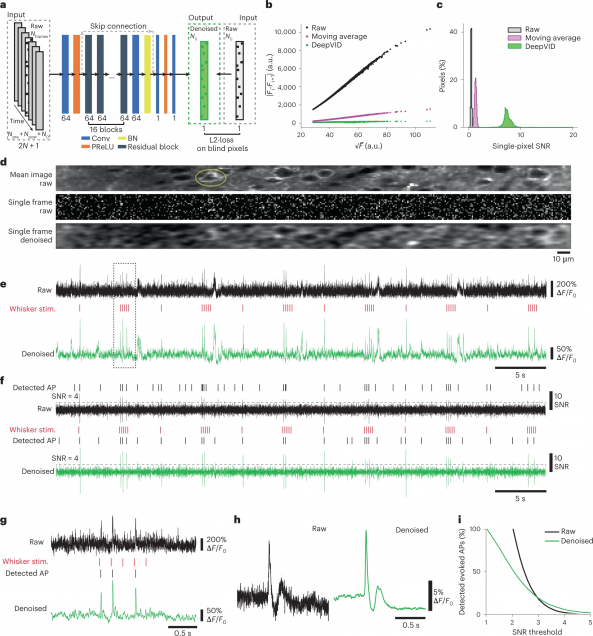
High-Speed Low-Light In Vivo Two-Photon Voltage Imaging of Large Neuronal Populations
Jelena Platisa, Xin Ye, Allison M Ahrens, Chang Liu, Ichun A Chen, Ian G Davison, Lei Tian, Vincent A Pieribone, Jerry L Chen
Nature Methods 20, 1095–1103 (2023).
⭑ Github Project
⭑ Spotlight: AI to the rescue of voltage imaging, Cell Reports Methods
Monitoring spiking activity across large neuronal populations at behaviorally relevant timescales is critical for understanding neural circuit function. Unlike calcium imaging, voltage imaging requires kilohertz sampling rates that reduce fluorescence detection to near shot-noise levels. High-photon flux excitation can overcome photon-limited shot noise, but photobleaching and photodamage restrict the number and duration of simultaneously imaged neurons. We investigated an alternative approach aimed at low two-photon flux, which is voltage imaging below the shot-noise limit. This framework involved developing positive-going voltage indicators with improved spike detection (SpikeyGi and SpikeyGi2); a two-photon microscope (‘SMURF’) for kilohertz frame rate imaging across a 0.4 mm × 0.4 mm field of view; and a self-supervised denoising algorithm (DeepVID) for inferring fluorescence from shot-noise-limited signals. Through these combined advances, we achieved simultaneous high-speed deep-tissue imaging of more than 100 densely labeled neurons over 1 hour in awake behaving mice. This demonstrates a scalable approach for voltage imaging across increasing neuronal populations.
Multiple-scattering simulator-trained neural network for intensity diffraction tomography
A. Matlock, J. Zhu, L. Tian
Optics Express 31, 4094-4107 (2023)
Recovering 3D phase features of complex biological samples traditionally sacrifices computational efficiency and processing time for physical model accuracy and reconstruction quality. Here, we overcome this challenge using an approximant-guided deep learning framework in a high-speed intensity diffraction tomography system. Applying a physics model simulator-based learning strategy trained entirely on natural image datasets, we show our network can robustly reconstruct complex 3D biological samples. To achieve highly efficient training and prediction, we implement a lightweight 2D network structure that utilizes a multi-channel input for encoding the axial information. We demonstrate this framework on experimental measurements of weakly scattering epithelial buccal cells and strongly scattering C. elegans worms. We benchmark the network’s performance against a state-of-the-art multiple-scattering model-based iterative reconstruction algorithm. We highlight the network’s robustness by reconstructing dynamic samples from a living worm video. We further emphasize the network’s generalization capabilities by recovering algae samples imaged from different experimental setups. To assess the prediction quality, we develop a quantitative evaluation metric to show that our predictions are consistent with both multiple-scattering physics and experimental measurements.
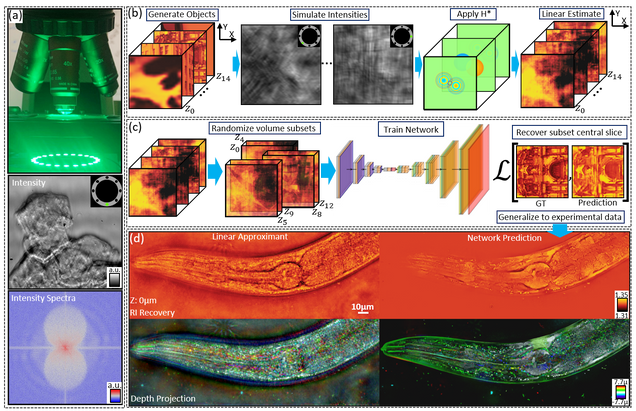
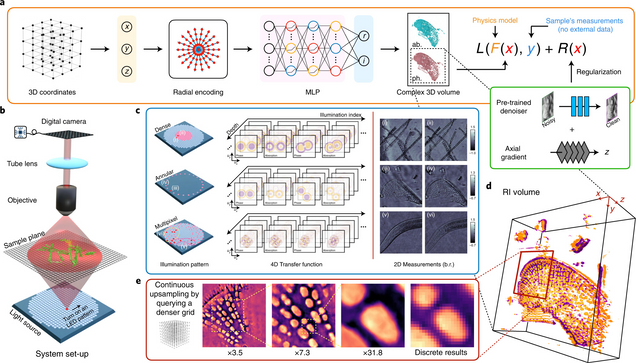
Recovery of Continuous 3D Refractive Index Maps from Discrete Intensity-Only Measurements using Neural Fields
Renhao Liu, Yu Sun, Jiabei Zhu, Lei Tian, Ulugbek Kamilov
Nature Machine Intelligence 4, 781–791 (2022).
Intensity diffraction tomography (IDT) refers to a class of optical microscopy techniques for imaging the three-dimensional refractive index (RI) distribution of a sample from a set of two-dimensional intensity-only measurements. The reconstruction of artefact-free RI maps is a fundamental challenge in IDT due to the loss of phase information and the missing-cone problem. Neural fields has recently emerged as a new deep learning approach for learning continuous representations of physical fields. The technique uses a coordinate-based neural network to represent the field by mapping the spatial coordinates to the corresponding physical quantities, in our case the complex-valued refractive index values. We present Deep Continuous Artefact-free RI Field (DeCAF) as a neural-fields-based IDT method that can learn a high-quality continuous representation of a RI volume from its intensity-only and limited-angle measurements. The representation in DeCAF is learned directly from the measurements of the test sample by using the IDT forward model without any ground-truth RI maps. We qualitatively and quantitatively evaluate DeCAF on the simulated and experimental biological samples. Our results show that DeCAF can generate high-contrast and artefact-free RI maps and lead to an up to 2.1-fold reduction in the mean squared error over existing methods.
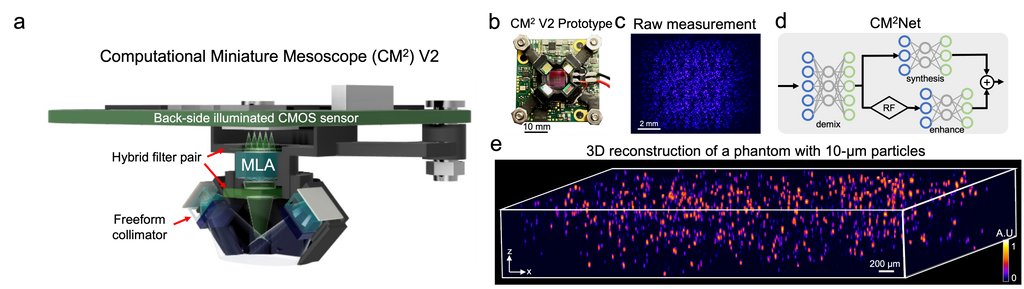
Deep learning-augmented Computational Miniature Mesoscope
Yujia Xue, Qianwan Yang, Guorong Hu, Kehan Guo, Lei Tian
Optica 9, 1009-1021 (2022)
Fluorescence microscopy is essential to study biological structures and dynamics. However, existing systems suffer from a tradeoff between field-of-view (FOV), resolution, and complexity, and thus cannot fulfill the emerging need of miniaturized platforms providing micron-scale resolution across centimeter-scale FOVs. To overcome this challenge, we developed Computational Miniature Mesoscope (CM
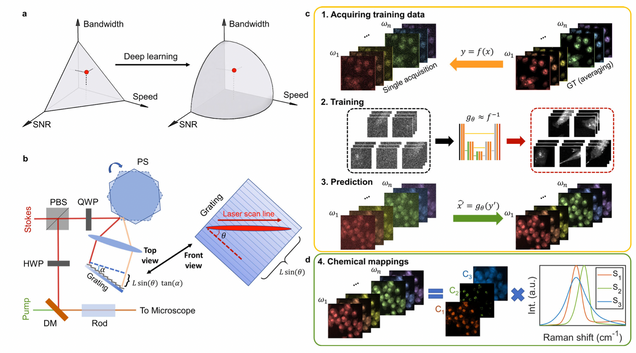
Microsecond fingerprint stimulated Raman spectroscopic imaging by ultrafast tuning and spatial-spectral learning
H. Lin, H.J. Lee, N. Tague, J.-B. Lugagne, C. Zong, F. Deng, J. Shin, L. Tian, W. Wong, M.J. Dunlop, J.-X. Cheng
Nature Communications 12(1) (2021).
Label-free vibrational imaging by stimulated Raman scattering (SRS) provides unprecedented insight into real-time chemical distributions. Specifically, SRS in the fingerprint region (400–1800 cm−1) can resolve multiple chemicals in a complex bio-environment. However, due to the intrinsic weak Raman cross-sections and the lack of ultrafast spectral acquisition schemes with high spectral fidelity, SRS in the fingerprint region is not viable for studying living cells or large-scale tissue samples. Here, we report a fingerprint spectroscopic SRS platform that acquires a distortion-free SRS spectrum at 10 cm−1 spectral resolution within 20 μs using a polygon scanner. Meanwhile, we significantly improve the signal-to-noise ratio by employing a spatial-spectral residual learning network, reaching a level comparable to that with 100 times integration. Collectively, our system enables high-speed vibrational spectroscopic imaging of multiple biomolecules in samples ranging from a single live microbe to a tissue slice.
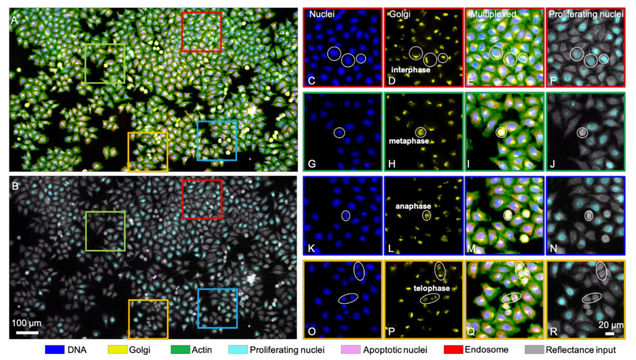
Single-cell cytometry via multiplexed fluorescence prediction by label-free reflectance microscopy
Shiyi Cheng, Sipei Fu, Yumi Mun Kim, Weiye Song, Yunzhe Li, Yujia Xue, Ji Yi, Lei Tian
Science Advances 15 Jan 2021: Vol. 7, no. 3, eabe0431
Traditional imaging cytometry uses fluorescence markers to identify specific structures but is limited in throughput by the labeling process. We develop a label-free technique that alleviates the physical staining and provides multiplexed readouts via a deep learning–augmented digital labeling method. We leverage the rich structural information and superior sensitivity in reflectance microscopy and show that digital labeling predicts accurate subcellular features after training on immunofluorescence images. We demonstrate up to three times improvement in the prediction accuracy over the state of the art. Beyond fluorescence prediction, we demonstrate that single cell–level structural phenotypes of cell cycles are correctly reproduced by the digital multiplexed images, including Golgi twins, Golgi haze during mitosis, and DNA synthesis. We further show that the multiplexed readouts enable accurate multiparametric single-cell profiling across a large cell population. Our method can markedly improve the throughput for imaging cytometry toward applications for phenotyping, pathology, and high-content screening.
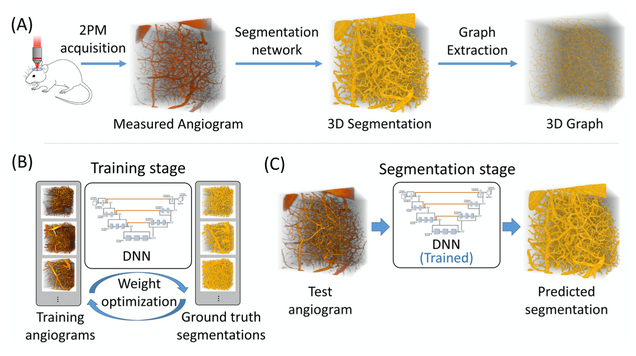
Anatomical modeling of brain vasculature in two-photon microscopy by generalizable deep learning
BME Frontiers, vol. 2021, Article ID 8620932
Segmentation of blood vessels from two-photon microscopy (2PM) angiograms of brains has important applications in hemodynamic analysis and disease diagnosis. Here we develop a generalizable deep-learning technique for accurate 2PM vascular segmentation of sizable regions in mouse brains acquired from multiple 2PM setups. In addition, the technique is computationally efficient, making it ideal for large-scale neurovascular analysis.
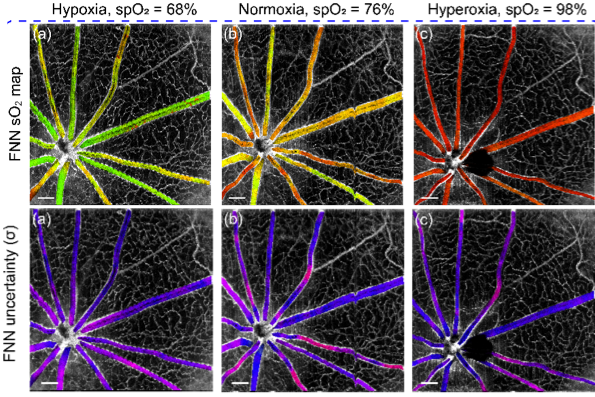
Deep spectral learning for label-free optical imaging oximetry with uncertainty quantification
, , ,
Measurement of blood oxygen saturation (sO2) by optical imaging oximetry provides invaluable insight into local tissue functions and metabolism. Despite different embodiments and modalities, all label-free optical-imaging oximetry techniques utilize the same principle of sO2-dependent spectral contrast from haemoglobin. Traditional approaches for quantifying sO2 often rely on analytical models that are fitted by the spectral measurements. These approaches in practice suffer from uncertainties due to biological variability, tissue geometry, light scattering, systemic spectral bias, and variations in the experimental conditions. Here, we propose a new data-driven approach, termed deep spectral learning (DSL), to achieve oximetry that is highly robust to experimental variations and, more importantly, able to provide uncertainty quantification for each sO2 prediction. To demonstrate the robustness and generalizability of DSL, we analyse data from two visible light optical coherence tomography (vis-OCT) setups across two separate in vivo experiments on rat retinas. Predictions made by DSL are highly adaptive to experimental variabilities as well as the depth-dependent backscattering spectra. Two neural-network-based models are tested and compared with the traditional least-squares fitting (LSF) method. The DSL-predicted sO2 shows significantly lower mean-square errors than those of the LSF. For the first time, we have demonstrated en face maps of retinal oximetry along with a pixel-wise confidence assessment. Our DSL overcomes several limitations of traditional approaches and provides a more flexible, robust, and reliable deep learning approach for in vivo non-invasive label-free optical oximetry.
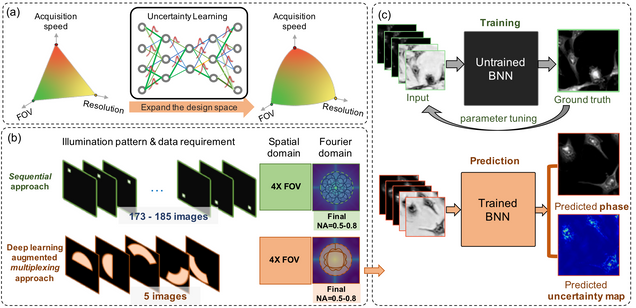
Reliable deep learning-based phase imaging with uncertainty quantification
Yujia Xue, Shiyi Cheng, Yunzhe Li, Lei Tian
Optica 6, 618-629 (2019).
Emerging deep-learning (DL)-based techniques have significant potential to revolutionize biomedical imaging. However, one outstanding challenge is the lack of reliability assessment in the DL predictions, whose errors are commonly revealed only in hindsight. Here, we propose a new Bayesian convolutional neural network (BNN)-based framework that overcomes this issue by quantifying the uncertainty of DL predictions. Foremost, we show that BNN-predicted uncertainty maps provide surrogate estimates of the true error from the network model and measurement itself. The uncertainty maps characterize imperfections often unknown in real-world applications, such as noise, model error, incomplete training data, and out-of-distribution testing data. Quantifying this uncertainty provides a per-pixel estimate of the confidence level of the DL prediction as well as the quality of the model and data set. We demonstrate this framework in the application of large space–bandwidth product phase imaging using a physics-guided coded illumination scheme. From only five multiplexed illumination measurements, our BNN predicts gigapixel phase images in both static and dynamic biological samples with quantitative credibility assessment. Furthermore, we show that low-certainty regions can identify spatially and temporally rare biological phenomena. We believe our uncertainty learning framework is widely applicable to many DL-based biomedical imaging techniques for assessing the reliability of DL predictions.
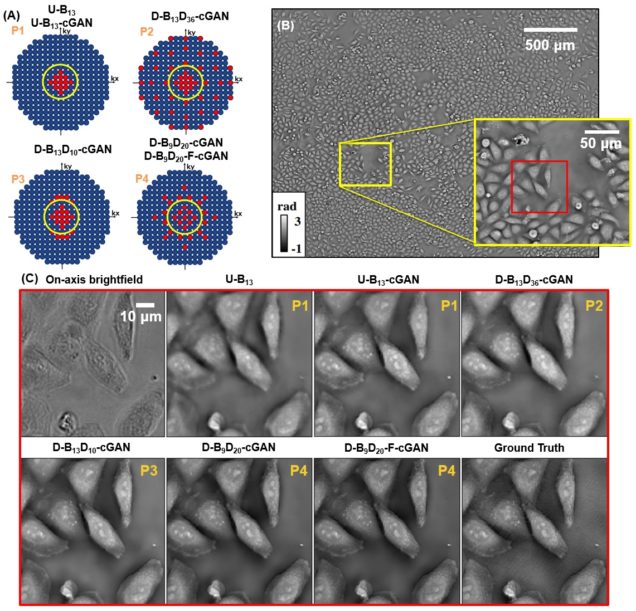
Deep learning approach to Fourier ptychographic microscopy
Thanh Nguyen, Yujia Xue, Yunzhe Li, Lei Tian, George Nehmetallah
Opt. Express 26, 26470-26484 (2018).
Convolutional neural networks (CNNs) have gained tremendous success in solving complex inverse problems. The aim of this work is to develop a novel CNN framework to reconstruct video sequence of dynamic live cells captured using a computational microscopy technique, Fourier ptychographic microscopy (FPM). The unique feature of the FPM is its capability to reconstruct images with both wide field-of-view (FOV) and high resolution, i.e. a large space-bandwidth-product (SBP), by taking a series of low resolution intensity images. For live cell imaging, a single FPM frame contains thousands of cell samples with different morphological features. Our idea is to fully exploit the statistical information provided by this large spatial ensemble so as to make predictions in a sequential measurement, without using any additional temporal dataset. Specifically, we show that it is possible to reconstruct high-SBP dynamic cell videos by a CNN trained only on the first FPM dataset captured at the beginning of a time-series experiment. Our CNN approach reconstructs a 12800X10800 pixels phase image using only ~25 seconds, a 50X speedup compared to the model-based FPM algorithm. In addition, the CNN further reduces the required number of images in each time frame by ~6X. Overall, this significantly improves the imaging throughput by reducing both the acquisition and computational times. The proposed CNN is based on the conditional generative adversarial network (cGAN) framework. Additionally, we also exploit transfer learning so that our pre-trained CNN can be further optimized to image other cell types. Our technique demonstrates a promising deep learning approach to continuously monitor large live-cell populations over an extended time and gather useful spatial and temporal information with sub-cellular resolution.