Publications
Full publication list at Google Scholar.

JOURNAL PUBLICATIONS
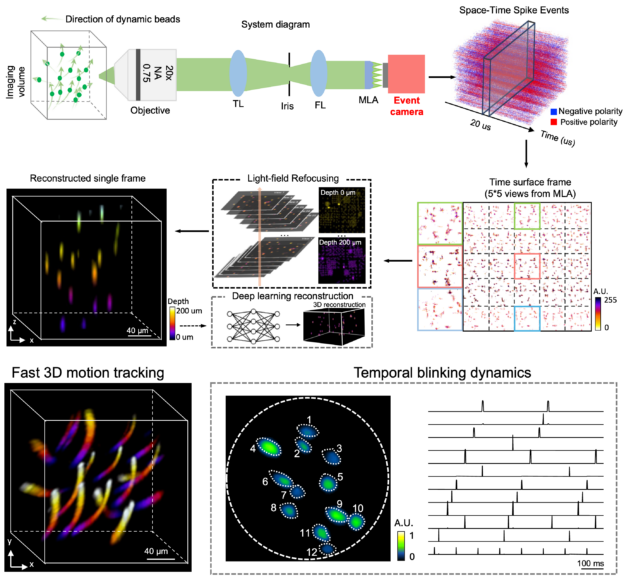
EventLFM: event camera integrated Fourier light field microscopy for ultrafast 3D imaging
Ruipeng Guo, Qianwan Yang, Andrew S. Chang, Guorong Hu, Joseph Greene, Christopher V. Gabel, Sixian You & Lei Tian
Light: Science & Applications 13: 144 (2024).
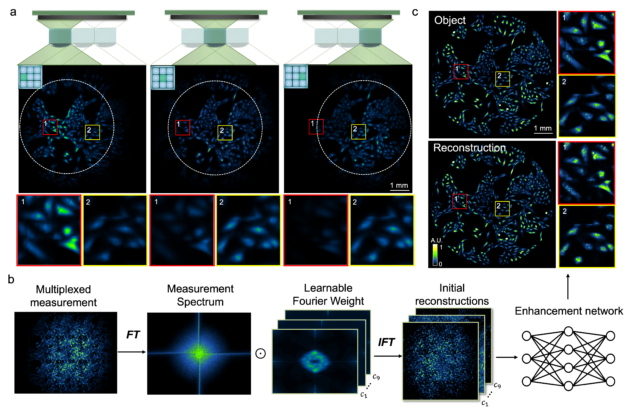
Wide-field, high-resolution reconstruction in computational multi-aperture miniscope using a Fourier neural network
Qianwan Yang, Ruipeng Guo, Guorong Hu, Yujia Xue, Yunzhe Li, and Lei Tian
Optica Vol. 11, Issue 6, pp. 860-871 (2024).
⭑ Github Project
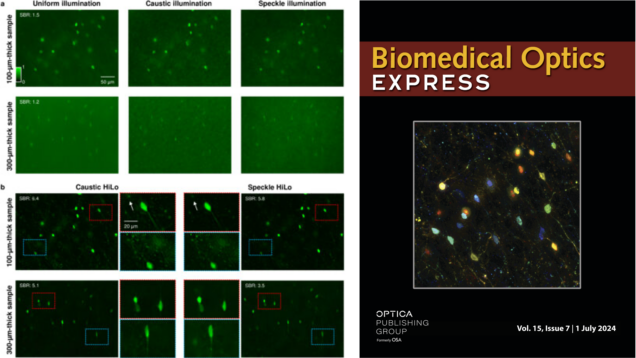
HiLo microscopy with caustic illumination
Guorong Hu, Joseph Greene, Jiabei Zhu, Qianwan Yang, Shuqi Zheng, Yunzhe Li, Jeffrey Alido, Ruipeng Guo, Jerome Mertz, and Lei Tian
Biomedical Optics Express Vol. 15, Issue 7, pp. 4101-4110 (2024). ⭑ On the Cover
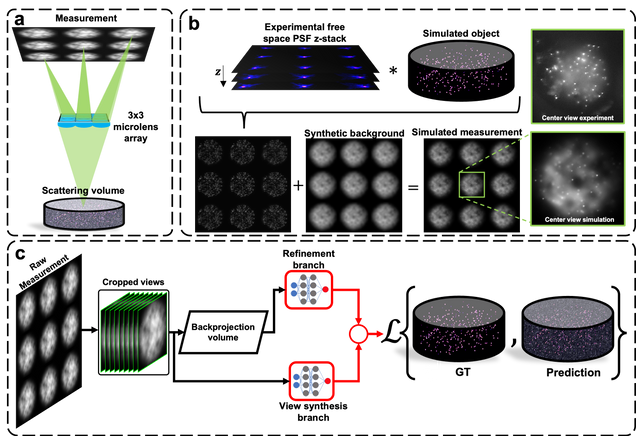
Robust single-shot 3D fluorescence imaging in scattering media with a simulator-trained neural network
J. Alido, J. Greene, Y. Xue, G. Hu, Y. Li, K. Monk, B. DeBenedicts, I. Davison, L. Tian
Optics Express Vol. 32, Issue 4, pp. 6241-6257 (2024).
⭑ Github Project
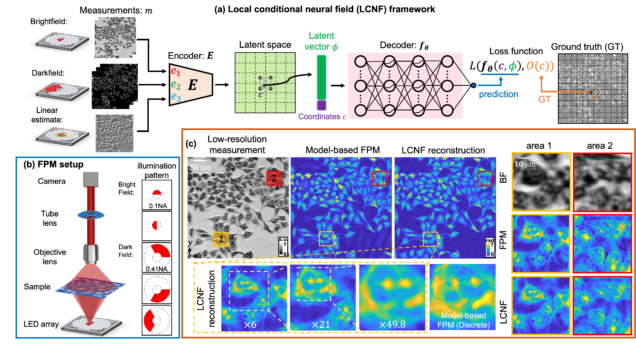
Local Conditional Neural Fields for Versatile and Generalizable Large-Scale Reconstructions in Computational Imaging
Hao Wang, Jiabei Zhu, Yunzhe Li, Qianwan Yang, Lei Tian
arXiv:2307.06207
⭑ Github Project
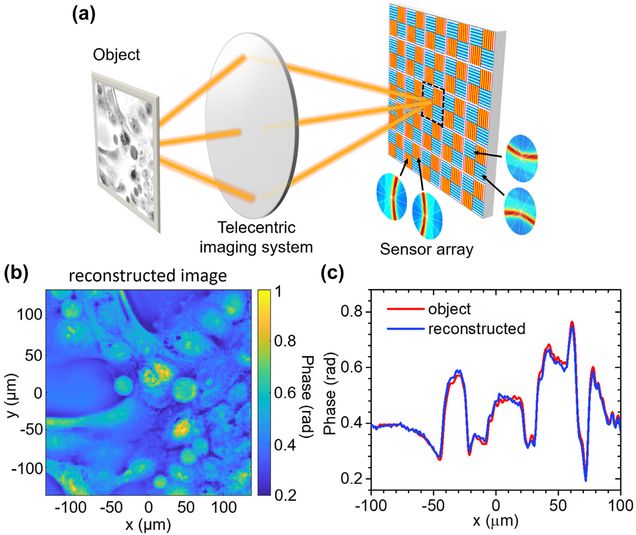
Asymmetric metasurface photodetectors for single-shot quantitative phase imaging
Jianing Liu , Hao Wang , Yuyu Li , Lei Tian, Roberto Paiella
Nanophotonics (2023).
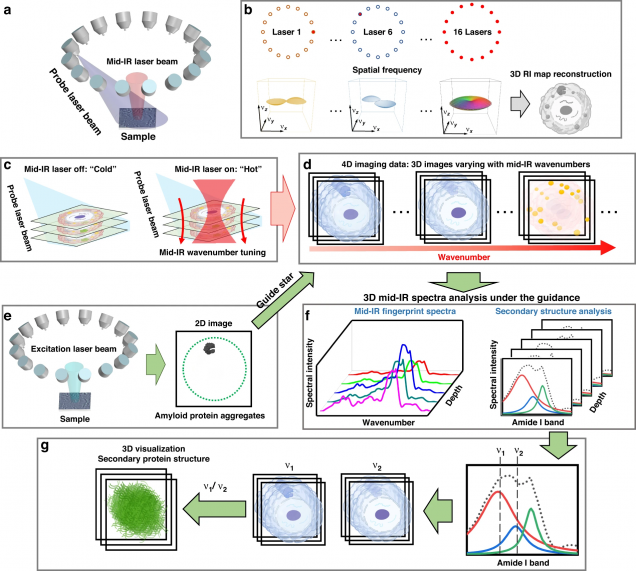
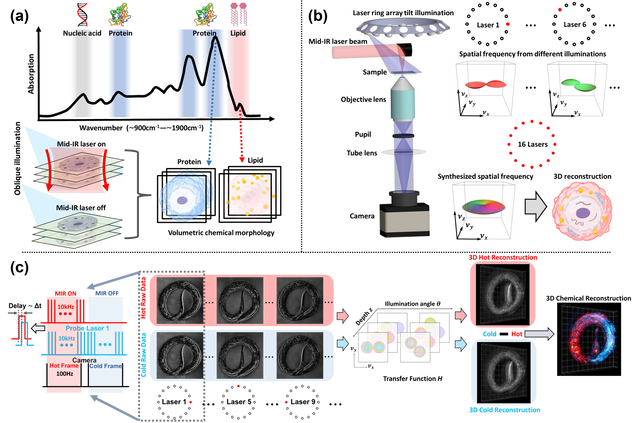
Mid-infrared chemical imaging of intracellular tau fibrils using fluorescence-guided computational photothermal microscopy
Jian Zhao, Lulu Jiang, Alex Matlock, Yihong Xu, Jiabei Zhu, Hongbo Zhu, Lei Tian, Benjamin Wolozin & Ji-Xin Cheng
Light: Science & Applications 12, 147 (2023).
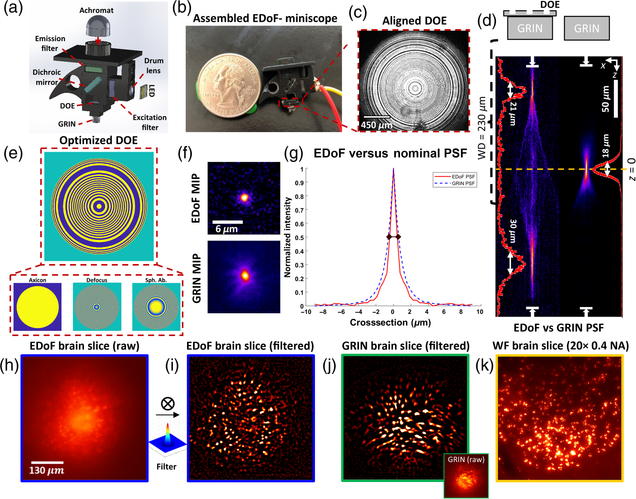
Pupil engineering for extended depth-of-field imaging in a fluorescence miniscope
Joseph Greene, Yujia Xue, Jeffrey Alido, Alex Matlock, Guorong Hu, Kivilcim Kiliç, Ian Davison, Lei Tian
Neurophotonics, Vol. 10, Issue 4, 044302 (2023).
High-Speed Low-Light In Vivo Two-Photon Voltage Imaging of Large Neuronal Populations
Jelena Platisa, Xin Ye, Allison M Ahrens, Chang Liu, Ichun A Chen, Ian G Davison, Lei Tian, Vincent A Pieribone, Jerry L Chen
Nature Methods 20, 1095–1103 (2023).
⭑ Github Project
⭑ Spotlight: AI to the rescue of voltage imaging, Cell Reports Methods
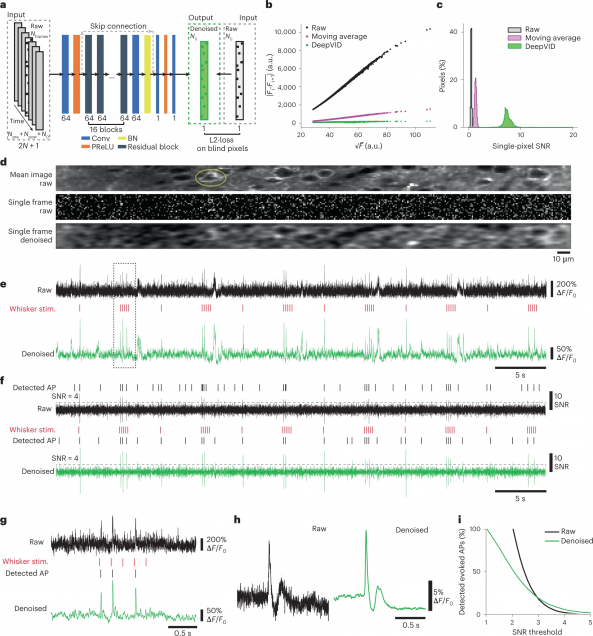
3D Chemical Imaging by Fluorescence-detected Mid-Infrared Photothermal Fourier Light Field Microscopy
Danchen Jia, Yi Zhang, Qianwan Yang, Yujia Xue, Yuying Tan, Zhongyue Guo, Meng Zhang, Lei Tian, Ji-Xin Cheng
Chem. Biomed. Imaging 2023, 1, 3, 260–267
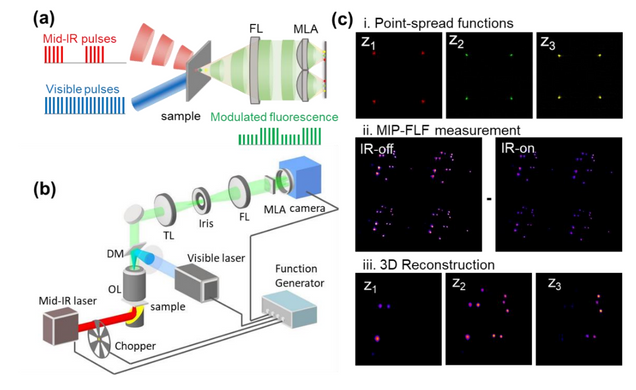
Fourier ptychographic topography
H. Wang, J. Zhu, J. Sung, G. Hu, J. Greene, Y. Li, S. Park, W. Kim, M. Lee, Y. Yang, L. Tian
Optics Express 31, pp. 11007-11018 (2023)
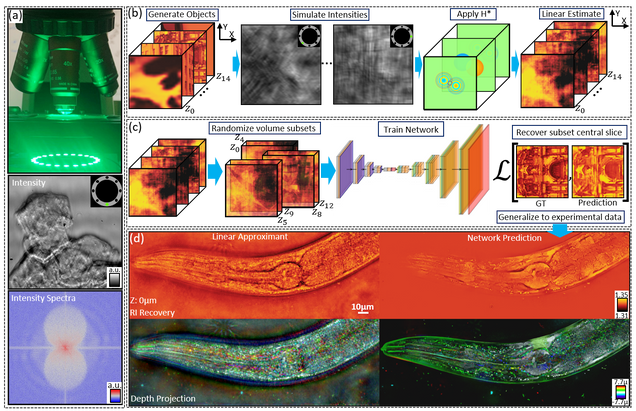
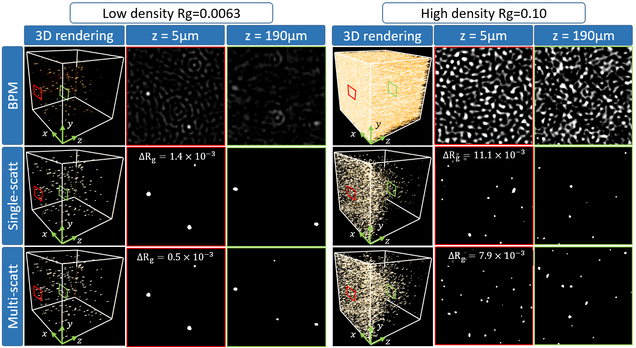
Multiple-scattering simulator-trained neural network for intensity diffraction tomography
A. Matlock, J. Zhu, L. Tian
Optics Express 31, 4094-4107 (2023)
Bond-Selective Intensity Diffraction Tomography
Jian Zhao, Alex Matlock, Hongbo Zhu, Ziqi Song, Jiabei Zhu, Biao Wang, Fukai Chen, Yuewei Zhan, Zhicong Chen, Yihong Xu, Xingchen Lin, Lei Tian, Ji-Xin Cheng
Nature Commun. 13, 7767 (2022).
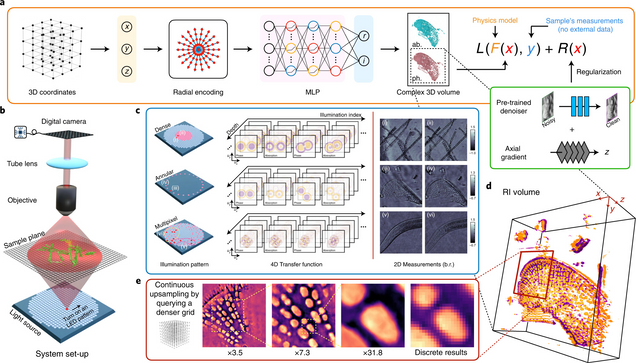
Recovery of Continuous 3D Refractive Index Maps from Discrete Intensity-Only Measurements using Neural Fields
Renhao Liu, Yu Sun, Jiabei Zhu, Lei Tian, Ulugbek Kamilov
Nature Machine Intelligence 4, 781–791 (2022).
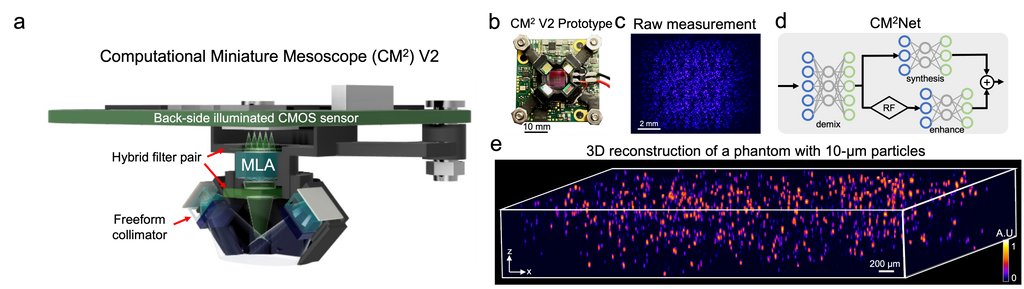
Deep-learning-augmented Computational Miniature Mesoscope
Yujia Xue, Qianwan Yang, Guorong Hu, Kehan Guo, Lei Tian
Optica 9, 1009-1021 (2022)
High-fidelity intensity diffraction tomography with a non-paraxial multiple-scattering model
Jiabei Zhu, Hao Wang, Lei Tian
Optics Express Vol. 30, Issue 18, pp. 32808-32821 (2022).
Optical spatial filtering with plasmonic directional image sensors
Jianing Liu, Hao Wang, Leonard C. Kogos, Yuyu Li, Yunzhe Li, Lei Tian, and Roberto Paiella
Optics Express Vol. 30, Issue 16, pp. 29074-29087 (2022).
⭑ Editors’ pick
Neurophotonic tools for microscopic measurements and manipulation: status report
Ahmed Abdelfattah, Sapna Ahuja, Taner Akkin, Srinivasa Rao Allu, David A. Boas, Joshua Brake, Erin M. Buckley, Robert E. Campbell, Anderson I. Chen, Xiaojun Cheng, Tomáš Cižmár, Irene Costantini, Massimo De Vittorio, Anna Devor, Patrick R. Doran, Mirna El Khatib, Valentina Emiliani, Natalie Fomin-Thunemann, Yeshaiahu Fainman, Tomás Fernández Alfonso, Christopher G. L. Ferri, Ariel Gilad, Xue Han, Andrew Harris, Elizabeth M. C. Hillman, Ute Hochgeschwender, Matthew G. Holt, Na Ji, Kivilcim Kiliç, Evelyn M. R. Lake, Lei Li, Tianqi Li, Philipp Mächler, Rickson C. Mesquita, Evan W. Miller, K.M. Naga Srinivas Nadella, U. Valentin Nägerl, Yusuke Nasu, Axel Nimmerjahn, Petra Ondrácková, Francesco S. Pavone, Citlali Perez Campos, Darcy S. Peterka, Filippo Pisano, Ferruccio Pisanello, Francesca Puppo, Bernardo L. Sabatini, Sanaz Sadegh, Sava Sakadžic, Shy Shoham, Sanaya N. Shroff, R. Angus Silver, Ruth R. Sims, Spencer L. Smith, Vivek J. Srinivasan, Martin Thunemann, Lei Tian, Lin Tian, Thomas Troxler, Antoine Valera, Alipasha Vaziri, Sergei A. Vinogradov, Flavia Vitale, Lihong V. Wang, Hana Uhlířová, Chris Xu, Changhuei Yang, Mu-Han Yang, Gary Yellen, Ofer Yizhar, Yongxin Zhao
Neurophotonics, 9(S1), 013001 (2022).

Adaptive 3D descattering with a dynamic synthesis network
Waleed Tahir, Hao Wang, Lei Tian
Light: Science & Applications 11, 42, 2022.
⭑ On the Cover
Roadmap on digital holography
Bahram Javidi, Artur Carnicer, Arun Anand, George Barbastathis, Wen Chen, Pietro Ferraro, J. W. Goodman, Ryoichi Horisaki, Kedar Khare, Malgorzata Kujawinska, Rainer A. Leitgeb, Pierre Marquet, Takanori Nomura, Aydogan Ozcan, YongKeun Park, Giancarlo Pedrini, Pascal Picart, Joseph Rosen, Genaro Saavedra, Natan T. Shaked, Adrian Stern, Enrique Tajahuerce, Lei Tian, Gordon Wetzstein, and Masahiro Yamaguchi
Optics Express Vol. 29, Issue 22, pp. 35078-35118 (2021).
Review of bio-optical imaging systems with a high space-bandwidth product
Jongchan Park, David J. Brady, Guoan Zheng, Lei Tian, Liang Gao
Advanced Photonics, 3(4), 044001 (2021).

Acousto-optic ptychography
M. Rosenfeld, G. Weinberg, D. Doktofsky, Y. Li, L. Tian, O. Katz
Optica 8, 936-943 (2021).
Microsecond fingerprint stimulated Raman spectroscopic imaging by ultrafast tuning and spatial-spectral learning
H. Lin, H.J. Lee, N. Tague, J.-B. Lugagne, C. Zong, F. Deng, J. Shin, L. Tian, W. Wong, M.J. Dunlop, J.-X. Cheng
Nature Communications 12(1) (2021).
⭑ In the news:
– BU ECE news.
Deep Learning in Biomedical Optics
L. Tian, B. Hunt, M. Bell, J. Yi, J. Smith, M. Ochoa, X. Intes, N. Durr
Lasers in Surgery and Medicine 53(6), 748, (2021).
⭑ On the Cover
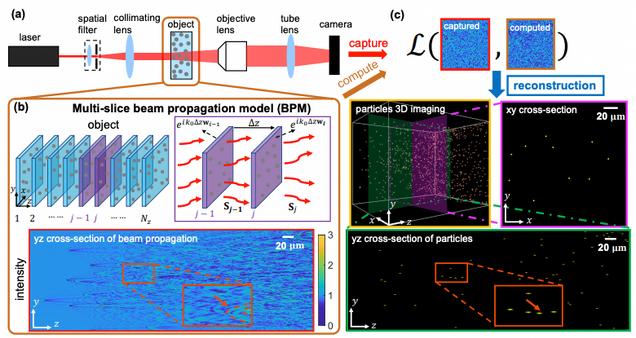
Large-scale holographic particle 3D imaging with the beam propagation model
Hao Wang, Waleed Tahir, Jiabei Zhu, Lei Tian
Opt. Express 29, 17159-17172 (2021)
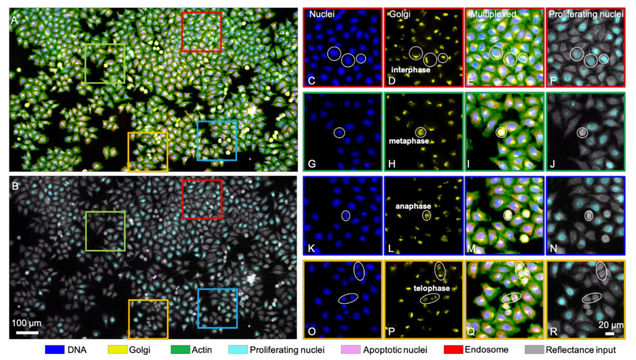
Single-cell cytometry via multiplexed fluorescence prediction by label-free reflectance microscopy
Shiyi Cheng, Sipei Fu, Yumi Mun Kim, Weiye Song, Yunzhe Li, Yujia Xue, Ji Yi, Lei Tian
Science Advances 15 Jan 2021: Vol. 7, no. 3, eabe0431
⭑ In the news:
– BU Hariri Institute: Deep Learning Allows for Digital Labeling of Multiple Cellular Structures
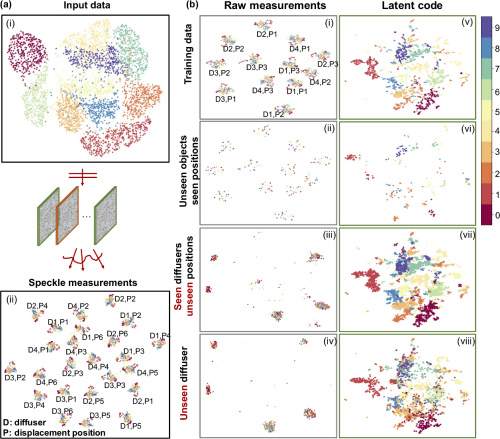
Displacement-agnostic coherent imaging through scatter with an interpretable deep neural network
Y Li, S Cheng, Y Xue, L Tian
Optics Express Vol. 29, Issue 2, pp. 2244-2257 (2021).
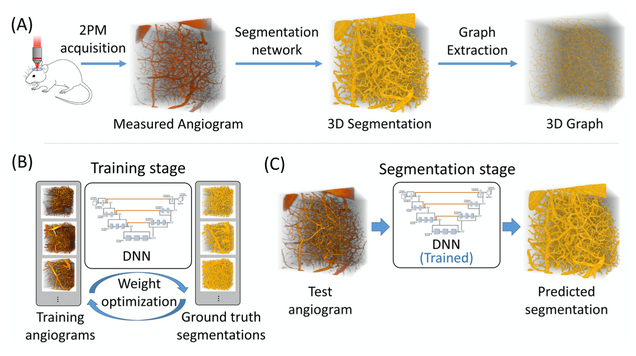
Anatomical modeling of brain vasculature in two-photon microscopy by generalizable deep learning
BME Frontiers, vol. 2021, Article ID 8620932
Single-Shot 3D Widefield Fluorescence Imaging with a Computational Miniature Mesoscope
Yujia Xue, Ian G. Davison, David A. Boas, Lei Tian
Science Advances 21 OCT 2020: EABB7508
⭑ On the Cover
⭑ In the news:
– BU ENG news: Brain Imaging Scaled Down
– BU CISE news: How Computational Imaging is Helping to Advance In-Vivo Studies of Brain Function
Single-Shot Ultraviolet Compressed Ultrafast Photography
Yingming Lai, Yujia Xue, Christian‐Yves Côté, Xianglei Liu, Antoine,Laramée, Nicolas Jaouen, François Légaré, Lei Tian, Jinyang Liang
Laser & Photonics Reviews 2020, 14, 2000122.
⭑ on the cover story

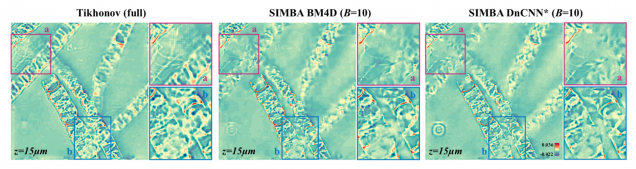
SIMBA: Scalable Inversion in Optical Tomography using Deep Denoising Priors
Zihui Wu, Yu Sun, Alex Matlock, Jiaming Liu, Lei Tian, Ulugbek S. Kamilov
IEEE Journal of Selected Topics in Signal Processing 14(6), 2020.
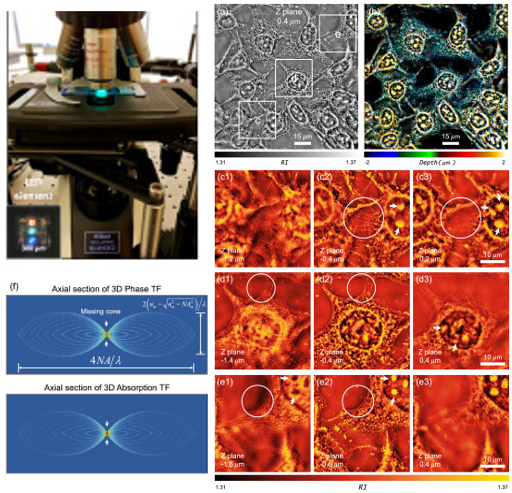
Resolution-enhanced intensity diffraction tomography in high numerical aperture label-free microscopy
Jiaji Li, Alex Matlock, Yunzhe Li, Qian Chen, Lei Tian, and Chao Zuo
Photonics Research 8(12), 1818-1826 (2020)
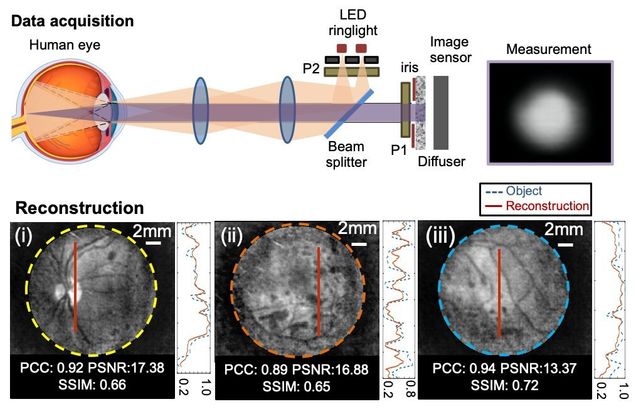
Diffuser-based computational imaging funduscope
Yunzhe Li, Gregory N. McKay, Nicholas J. Durr, Lei Tian
Optics Express 28, 19641-19654 (2020)
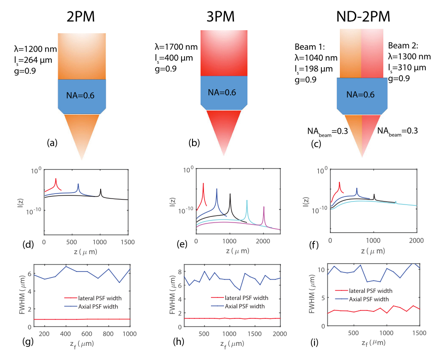
Comparing the fundamental imaging depth limit of two-photon, three-photon, and non-degenerate two-photon microscopy
Xiaojun Cheng, Sanaz Sadegh, Sharvari Zilpelwar, Anna Devor, Lei Tian, and David A. Boas
Optics Letters 45, pp. 2934-2937 (2020).
Plasmonic ommatidia for lensless compound-eye vision
Leonard C. Kogos, Yunzhe Li, Jianing Liu, Yuyu Li, Lei Tian & Roberto Paiella
Nature Communications 11: 1637 (2020).
⭑ In the news:
– BU ENG news: A Bug’s-Eye View
⭑ Highlighted in OPN Optics in 2020: Plasmonic Computational Compound-Eye Camera
High-Throughput, High-Resolution Interferometric Light Microscopy of Biological Nanoparticles
C. Yurdakul, O. Avci, A. Matlock, A. J Devaux, M. V Quintero, E. Ozbay, R. A Davey, J. H Connor, W C. Karl, L. Tian, M S. Ünlü
ACS Nano 2020, 14, 2, 2002-2013
LED array reflectance microscopy for scattering-based multi-contrast imaging
Weiye Song, Alex Matlock, Sipei Fu, Xiaodan Qin, Hui Feng, Christopher V. Gabel, Lei Tian, and Ji Yi
Opt. Lett. 45, 1647-1650 (2020)
Design of a high-resolution light field miniscope for volumetric imaging in scattering tissue
Yanqin Chen, Bo Xiong, Yujia Xue, Xin Jin, Joseph Greene, and Lei Tian
Biomedical Optics Express. 11, pp. 1662-1678 (2020).
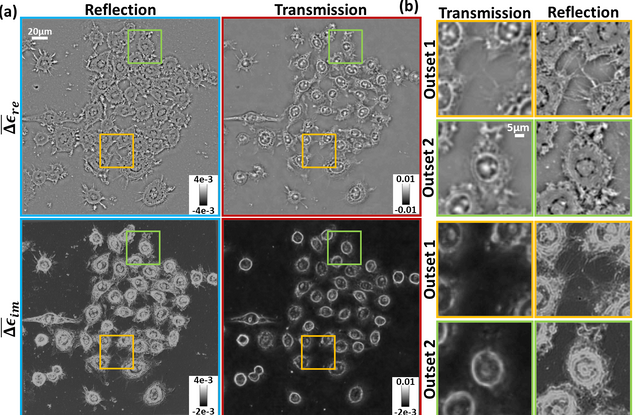
Inverse scattering for reflection intensity phase microscopy
Alex Matlock, Anne Sentenac, Patrick C. Chaumet, Ji Yi, and Lei Tian
Biomedical Optics Express. 11, pp. 911-926 (2020)
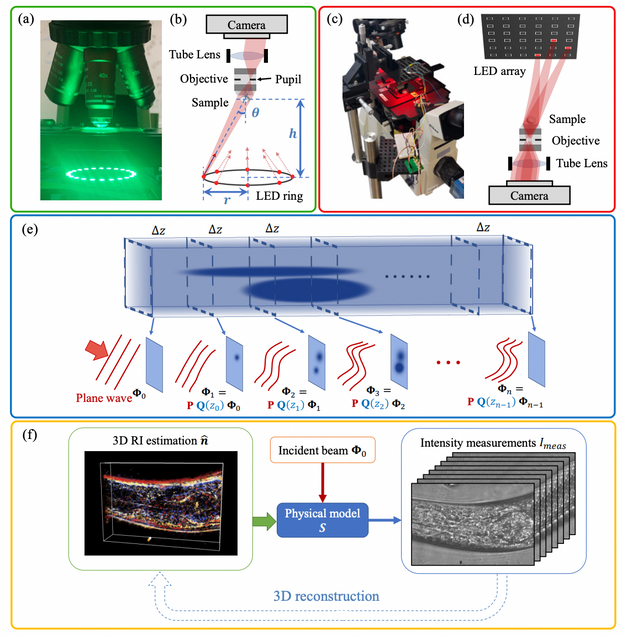
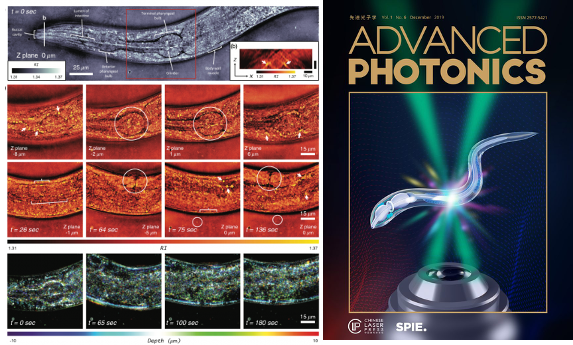
High-speed in vitro intensity diffraction tomography
Jiaji Li, Alex Matlock, Yunzhe Li, Qian Chen, Chao Zuo, Lei Tian
Advanced Photonics, 1(6), 066004 (2019).
⭑ on the cover story
⭑ Highlighted at Programmable LED ring enables label-free 3D tomography for conventional microscopes
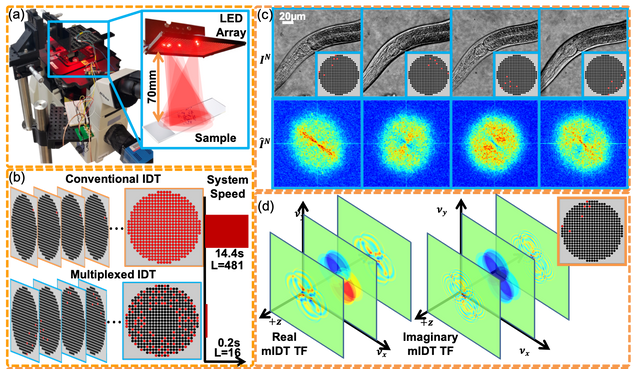
High-throughput, volumetric quantitative phase imaging with multiplexed intensity diffraction tomography
Alex Matlock, Lei Tian
Biomed. Opt. Express 10, pp. 6432-6448 (2019).
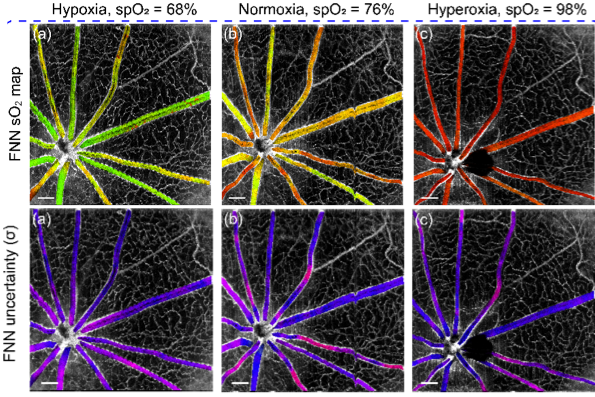
Deep spectral learning for label-free optical imaging oximetry with uncertainty quantification
, , ,
Development of a beam propagation method to simulate the point spread function degradation in scattering media
Xiaojun Cheng, Yunzhe Li, Jerome Mertz, Sava Sakadžić, Anna Devor, David A. Boas, Lei Tian
Opt. Lett. 44, 4989-4992 (2019).
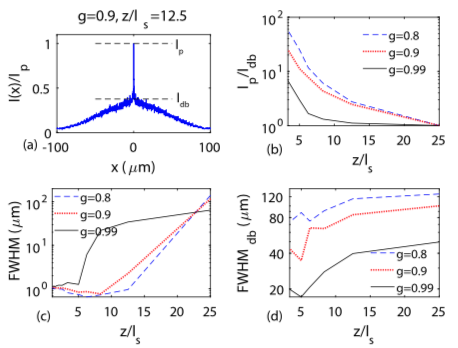
Holographic particle-localization under multiple scattering
Waleed Tahir, Ulugbek S. Kamilov, Lei Tian
Advanced Photonics, 1(3), 036003 (2019).
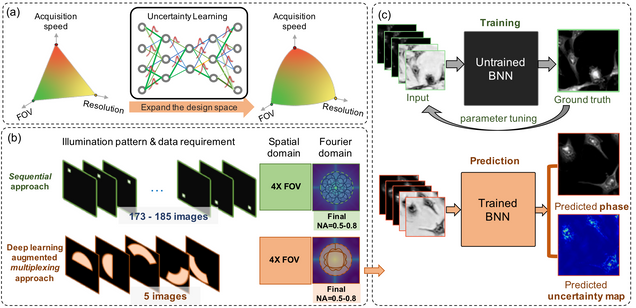
Reliable deep learning-based phase imaging with uncertainty quantification
Yujia Xue, Shiyi Cheng, Yunzhe Li, Lei Tian
Optica 6, 618-629 (2019).
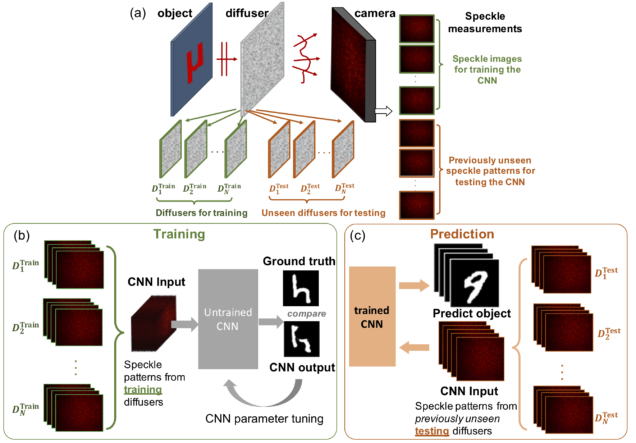
Deep speckle correlation: a deep learning approach towards scalable imaging through scattering media
Yunzhe Li, Yujia Xue, Lei Tian
Optica 5, 1181-1190 (2018).
⭑ Top 5 most cited articles in Optica published in 2018 (Source: Google Scholar)
Deep learning approach to Fourier ptychographic microscopy
Thanh Nguyen, Yujia Xue, Yunzhe Li, Lei Tian, George Nehmetallah
Opt. Express 26, 26470-26484 (2018).
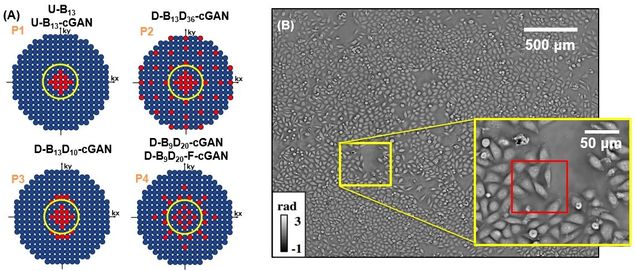
High-throughput intensity diffraction tomography with a computational microscope
Ruilong Ling, Waleed Tahir, Hsing-Ying Lin, Hakho Lee, and Lei Tian
Biomed. Opt. Express 9, 2130-2141 (2018)
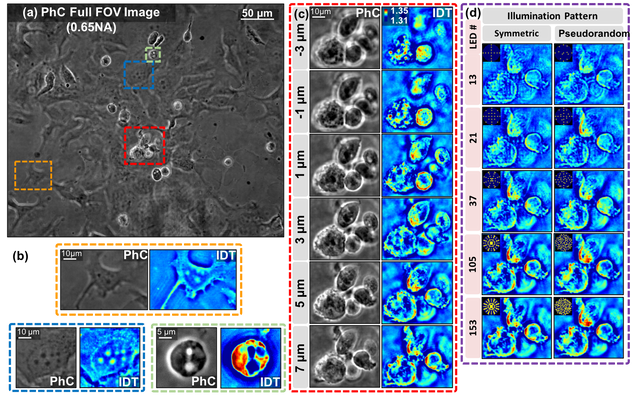
Structured illumination microscopy with unknown patterns and a statistical prior
Li-Hao Yeh, Lei Tian, and Laura Waller
Biomed. Opt. Express 8, 695-711 (2017).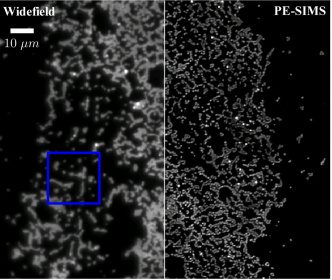
Compressive holographic video
Zihao Wang, Leonidas Spinoulas, Kuan He, Lei Tian, Oliver Cossairt, Aggelos K. Katsaggelos, and Huaijin Chen
Opt. Express 25, 250-262 (2017).
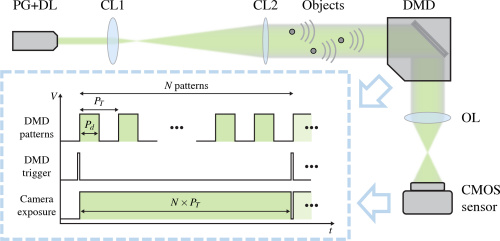
Nonlinear Optimization Algorithm for Partially Coherent Phase Retrieval and Source Recovery
J. Zhong, L. Tian, P. Varma, L. Waller
IEEE Transactions on Computational Imaging 2 (3), 310 – 322 (2016).
Computational illumination for high-speed in vitro Fourier ptychographic microscopy
L. Tian, Z. Liu, L. Yeh, M. Chen, J. Zhong, L. Waller
Optica 2(10), 904-911 (2015).
Computational imaging: Machine learning for 3D microscopy
L. Waller, L. Tian
Nature, 523, 416–417 (2015).
Quantitative differential phase contrast imaging in an LED array microscope
L. Tian, L. Waller
Opt. Express 23, 11394-11403 (2015).
3D intensity and phase imaging from light field measurements in an LED array microscope
Lei Tian, L. Waller
Optica 2, 104-111 (2015).
⭑ the 15 Most Cited Articles in Optica published in 2015 (Source: OSA, 2019)
Real-time brightfield, darkfield and phase contrast imaging in an LED array microscope
Z. Liu, Lei Tian, S. Liu, L. Waller
Journal of Biomedical Optics, 19(10), 106002 (2014).
![]()
Multiplexed coded illumination for Fourier Ptychography with an LED array microscope
Lei Tian, X. Li, K. Ramchandran, L. Waller
Biomedical Optics Express 5, 2376-2389 (2014).
⭑ the decade’s most highly cited Articles in Biomed. Opt. Express (Source: OSA, 2020)
⭑ Highly cited (Top 1%) papers between 2008-2018 (source: Web of Science, 2019)
3D differential phase contrast microscopy with computational illumination using an LED array
Lei Tian, J. Wang, L. Waller
Optics Letters 39, 1326 – 1329 (2014).