(2AI2BIO)* Group (Former Genomic Systems Biology Laboratory)
(2AI2BIO)*: Bridges from Artificial Intelligence to Understanding and Reprograming Biology
The * is a classic notation in Chomsky Grammars and means repeat — 2AI2BIO2AI2BIO…..
Imaginative science and collaborations produce magic, value and life long friendships
In applications of AI to life sciences and medicine, imagination is not sufficient because “Nature has the bigger imagination”. Humbling!
Imagination, humility and deep respect for structures and poorly understood but often magical (until discovery) rules allow us to make progress.
We focus on building bridges between AI and Biomedical Science / Engineering. Our focus is on new ideas that have a good chance to innovate in both AI and Biology/Biomedical Science/Engineering (rather than just using off the shelf AI prediction techniques on biological or clinical data). We call this bridge (2AI2BIO)* – a continuous bi-directional loop of innovation connecting AI and Biomedical Science and Engineering. We are also working on AI Safety in Biomedical Systems and Beyond.
We will continue consulting and collaborating in both core AI (ML/KR/etc) and Bioinformatics/Computational and Systems Biology.
We made two predictions in our 1993-5 papers in our AI Laboratory at Johns Hopkins University that still provide a vision for the utility of AI in reasoning about biology and modeling/understanding/reprogramming biological systems. We and others expanded on these predictions and implemented parts of this vision. However, we are far from completion, even with the revolutionary ML/Deep Learning advances and acceleration in the last 10 years. We enclose our main prediction from 1993.
“To summarize, scientific analysis of data is an important potential application of Artificial Intelligence (AI) research. We believe that the ultimate data analysis system using AI techniques will have a wide range of tools at its disposal and will adaptively choose various methods.
It will be able to generate simulations automatically and verify the model it constructed with the data generated during these simulations. When the model does not fit the observed results the system will try to explain the source of error, conduct additional experiments, and choose a different model by modifying system parameters. If it needs user assistance, it will produce a simple low-dimensional view of the constructed model and the data. This will allow the user to guide the system toward constructing a new model and/or generating the next set of experiments.”
Simon Kasif (Laboratory Director)
CV
Time Table of Significant Work
Selected Unusual Projects and Stories from Group Members
We are fully committed to deploying AI responsibly and safely in the service of health, wellness, justice, equity, fairness, reducing or eliminating suffering and intellectual diversity.
To dream is easy, to build is harder, to dream and build is the hardest
While we have deep and equal respect to all forms of innovation, we tend to focus on radical or early innovation and/or “extreme collaboration”. We are also committed to useful innovation and we often initiate or collaborate on projects that produce open access developments and other community service. Few selected examples of such collaborative innovative projects members of our group were fortunate to contribute to (in collaboration with truly off scale colleagues around the world) are listed below. We are not including solo work by the PI. Details of these projects and collaborators are available in the Time Table
-
The first parallel logic programming AI system that was actually implemented on a parallel machine (early 1980s). While it was “hanging on a wire”, it recognized the essential role of parallelism in AI and implemented in a rudimentary form a number of useful practical constructs. One of them was the use of the fork-join programming construct in AI systems. Much later, these early ideas were exponentially scaled up in the seminal work at Google by Jeff Dean et al in the system Map-Reduce.
-
The first scalable open access and widely used decision tree machine learning (AI) system using randomization (early 1990s)
-
An early proposal for Graphical AI Models for Biology (1992-)
-
Early and really rudimentary Generative AI methodologies for Computational Biology based on graphical models – a step towards using AI for Synthetic Biology and Protein Design (1993-)
-
Human-Centered AI systems initiative at NSF (mid 1990s).
-
Limited memory learning (among the very early papers that eventually led to formal models of streaming) .. 1990.
-
Learning with a Helpful Teacher (very early 1990-s).
-
Widely used and first whole genome microbial comparison and analysis system (mid 1990s)
-
Widely used microbial gene finder with exceptional accuracy using AI methods (mid-late 1990s).
-
Human Genome Project: Initial Draft (1999-)
-
Very early experimental validation of regulatory network predictions in human cancer cells.
-
Network propagation based gene function prediction using AI methods (early 2000-s)
-
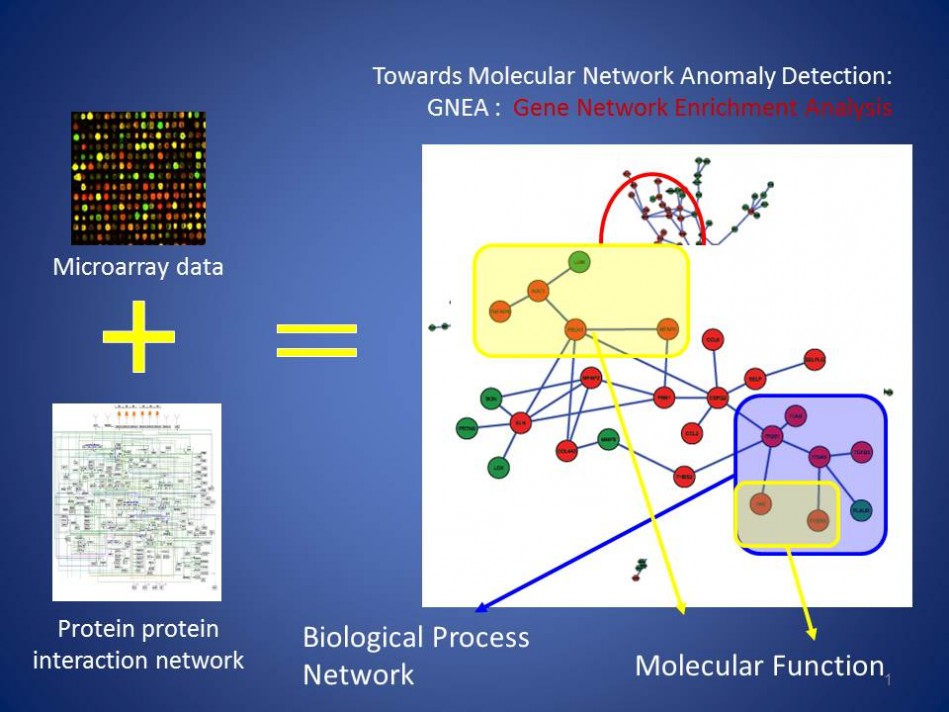
Network Based Anomaly Detection for Disease Identification with a focus on Metabolic Diseases, Diabetes and Aging (mid 2000)
-
AI based guidance of community experiments to test gene function : The COMBREX PROJECT (2003-)… a marriage of AI, Biology and Amazon Turk.
-
Biological Context Networks (2004-)
-
Combinatorial Experiment Design with Application to Liquid Biopsy (2002-)
-
One of the earliest large scale experimental validation of machine learning based regulatory network prediction in bacteria (2007).
-
One of the earliest Drug Side Effects Detection on Twitter using AI.
-
Facts matter! Tracing AI Predictions to Experimental Sources (2004, 2008-, 2020-)
-
Artificial Tikkun Olam POV (review and early analysis of AI opportunties and AI risks with some solutions 2018-)
-
Transforming Biology with AI (2020-)
-
APPs
-
AI Safety Initiatives (2004, 2008-, 2020-)
Anti-war Ukraine
Simon Kasif (Laboratory Director)
Time Table of Significant Work

Nature is sending another wake-up call. We must listen!
Principal Investigator – Selected Work Highlights and Lessons — in construction)
We love and appreciate all animals but our favorite “non-model organism” that we don’t study, just love and treat as family are dogs :
If we needed more proofs that a serious action is needed here is one.
More dogs pictures.

Ray and Whisk
Our work (AI2BIO)*
Our main focus is on application of Artificial Intelligence to human and bacterial genomic systems biology and medicine. But we studied (usually comparatively) many systems in model organisms such as worms (C.elegans), Flies (Drosophila), Yeast, Mouse, and other organisms. Evolution provides many clues and often direct answers. We support in spirit and work all the amazing and selfless work done by the Biomedical Databases Community.
We can think anywhere
Even during a pandemic (sober and inspiring writing from a front line doctor)
We strive to generate computational and biomedical ideas, concepts, techniques, technology platforms, systems, algorithms, software and thought frameworks to transform medicine and improve fundamental understanding of biological systems.
Artificial Intelligence methods drive our computational thinking about biology and biomedical methods.
Sometimes we succeed :)
We are one of the laboratories that contribute to the rich environment in the area of computational and systems biology at Boston University and multiple other institutions in the Boston area such as Joslin Diabetes, Center, MGH, Children’s Hospital (the MIT/Harvard Health Sciences and Technology Division) , the Broad Institute and others.
Before we describe our (2AI2BIO)* Genomic Systems Biology laboratory it is important to recognize the history of this area at Boston University and our small place in it.
You can find a brief description of this legacy here.
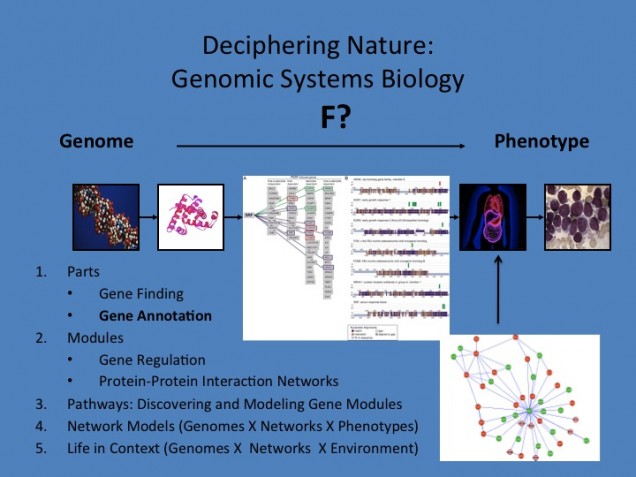
Genomic Systems Biology: 1) Can AI Decode and Program Nature? 2) Does AI need to understand Nature to pass the Turing Test?
Our primary long term interests are in human biology/physiology and bacteria. However, our group contributed to knowledge in multiple model organisms including Yeast, C.elegans, Drosophila and Mouse.
NETWORK SIGNATURES OF DISEASE
COmputational BRidges to EXperiments COMBREX (figure by Irina Glotova)