Research

1. Systems Biology of Aging and Longevity
Aging is a complex process that is controlled by a combination of genetic and environmental factors. A number of diverse genes have been shown to regulate aging. However, very little is known how different aging genes and pathways interact with each other. Using yeast Saccharomyces cerevisiae as a model, our laboratory employs systems biology approaches to build a regulatory network and define how aging pathways interact. For this, we utilize ribosome profiling and next-generation sequencing to quantitatively analyze genome-wide transcriptional and translational changes that occur in yeast cells with aging, in response to dietary restriction and genetic interventions known to extend lifespan. This information is integrated to develop interaction networks and identify key signaling pathways and regulators of protein translation that mediate the effects of interventions on aging.
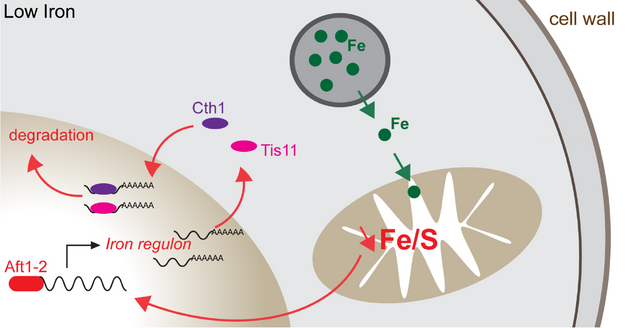
2. The Role of Iron Homeostasis in Aging
Iron is an essential trace element, which serves as a cofactor for enzymes involved in multiple metabolic pathways, including synthesis of DNA, ribosome biogenesis, lipid metabolism, and mitochondrial oxidative phosphorylation. In eukaryotes, iron utilization and storage are tightly controlled. Dysregulation of iron homeostasis has been implicated in the development of many human age-related diseases such as type 2 diabetes, cancer, neurodegeneration and cardiovascular disease. However, how iron homeostasis contributes to organismal aging remains unclear. We recently found that deficiency of Cth1 and Cth2, two mRNA-binding proteins involved in post-transcriptional regulation of iron homeostasis, leads to lifespan extension in yeast. Intriguingly, we discovered that aging in yeast is also accompanied by increased activity of the iron starvation response. In this project, we use a combination of sequencing and single-cell imaging methods to identify genes that are translationally regulated by Cth1 and Cth2 in response to iron deficiency, characterize metabolic consequences of iron dysregulation, and investigate the role of iron homeostasis genes in lifespan control. Given that genes regulated by iron are evolutionarily conserved among eukaryotes, these studies will significantly improve our understanding of the molecular mechanisms underlying the regulation of iron homeostasis and their role in aging.
3. Conserved Mechanisms of Adaptation to ER Stress
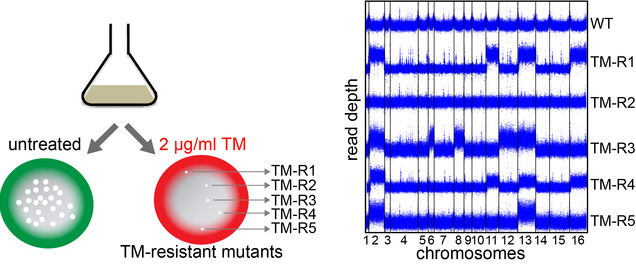
In eukaryotes, about one-third of all proteins are folded in the endoplasmic reticulum (ER). When protein folding demand exceeds capacity of the ER, the unfolded protein response (UPR) is activated to counteract ER stress. This signaling pathway initiates degradation of misfolded proteins, and leads to inhibition of translation and increased expression of chaperones and oxidative folding components that enhance ER protein folding capacity. However, the mechanisms of adaptation to ER stress are not restricted to the UPR. While the role of the UPR signaling in maintaining ER homeostasis has been extensively studied, there are still significant gaps in our understanding of the mechanisms by which cells adapt to accumulation of misfolded proteins in the ER. We recently performed a genetic screen in yeast to identify mutations that confer resistance to tunicamycin-induced ER stress. Excitingly, these studies uncovered aneuploidy as an important mediator of ER stress resistance, which protects cells against protein misfolding in a UPR-independent manner. The goal of this project is to characterize adaptive responses by which eukaryotic cells protect themselves from ER stress and investigate the link between metabolism and ER stress resistance. Building on our findings, we aim to address the following questions:
(1) What are the key UPR-independent mechanisms that eukaryotic cells use to counteract ER stress?
(2) How do metabolic adaptations mediated by the UPR contribute to the resolution of protein misfolding and restoration of protein homeostasis?
(3) How do the UPR and UPR-independent mechanisms for ER stress resistance dynamically change at the single-cell level?