Research
Bioimaging & Informatics Laboratory (BIL)
The goal of our lab is to better understand mechanisms underlying brain aging and explore biological pathways vital for increasing brain health-span using cutting-edge bioimaging and data science approaches. Much of our recent work focuses on developing imaging biomarkers, designing complex biological network models that combine datasets from multiple domains, and bridging the gap between laboratory neuroscience and human population neuroscience using imaging techniques and biomedical data fusion methods available for both animal models (for rodents and non-human primates) and humans. Students and research associates who train in our lab leave with a broad education in the latest in-vivo bioimaging techniques (e.g., non-invasive Blood-Brain Barrier imaging, MRI of neuro-inflammation, Brain chemical imaging, glial PET imaging, etc.), data science (e.g., Machine Learning, Deep Learning, Bayesian network, etc.), 3D histology, analysis framework for Radio-genomics/transcriptomics, etc.), along with skills in leadership, communication, and grant preparation. We also have several long-term national & international collaboration projects in which our students actively participate.
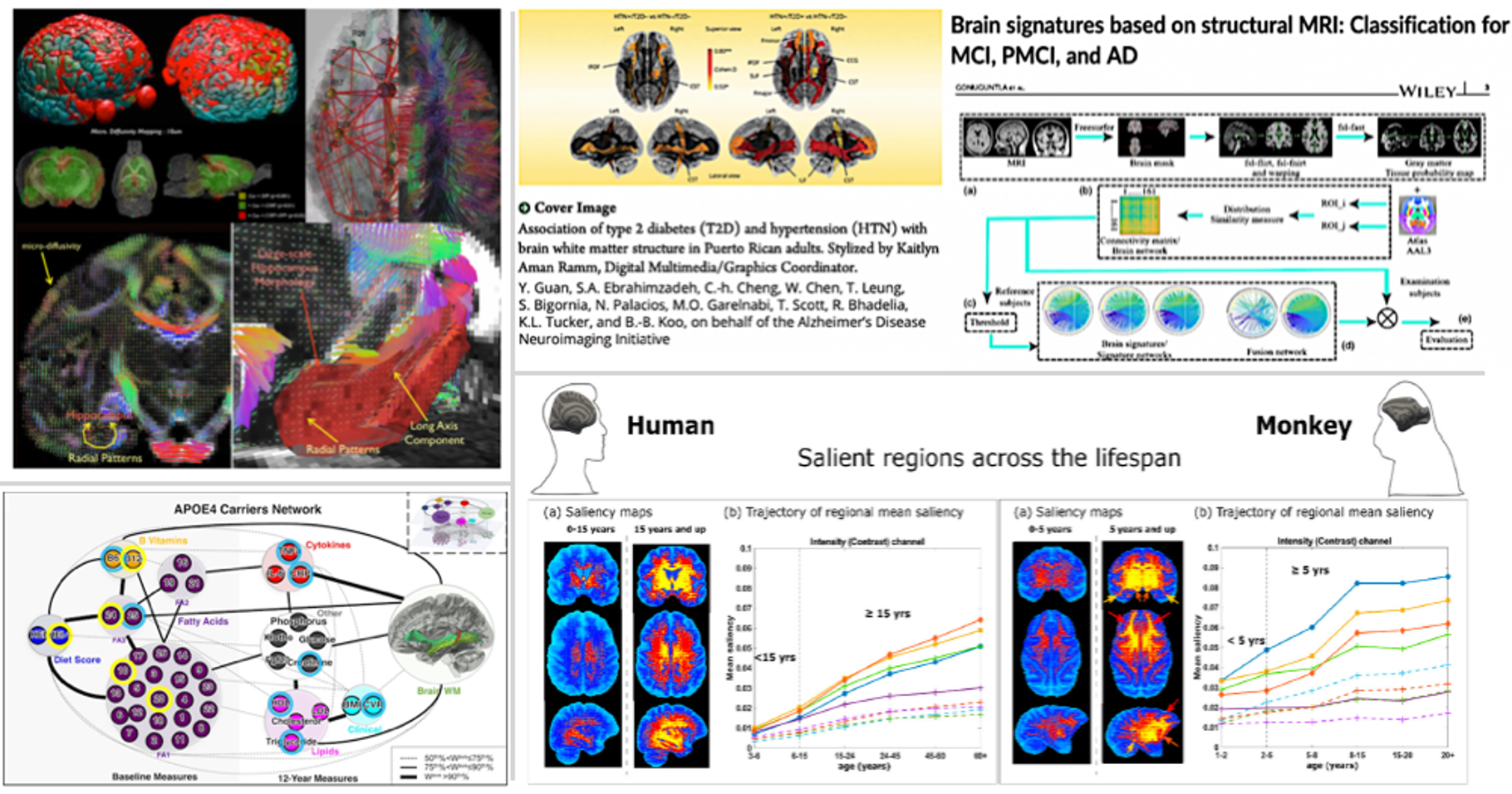
The tissue water diffusion information can be potentially sensitive to many factors including axons, dendrites, as well as myelinated fibers. Recent studies based on diffusion MRI have shown that the diffusion imaging can provide a sensitive measurement to astroglial plasticity induced by a learning task or mild traumatic brain injury. We have been working on developing high-order diffusion magnetic resonance imaging framework (hd-MRIf) to reconstruct brain water diffusivity information with improved sensitivities. The hd-MRIf was applied to assess brain microstructures and also, demonstrated a record of successful applications of hd-MRIf in in-vivo imaging works.
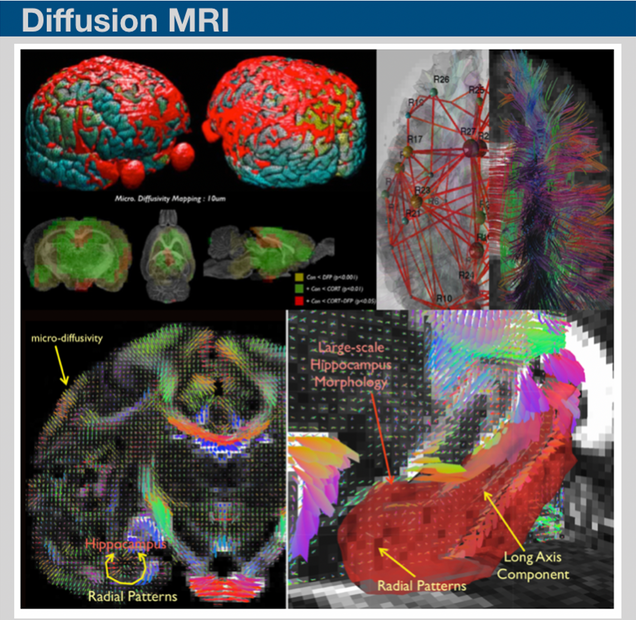
Resting-state fluctuations in the blood oxygenation level-dependent signal from functional MRI may provide novel insights on the basis of neural functional architecture. Recent techniques on assessing functional connectivity analysis have been providing valuable data resources for discriminating the mild disease progression symptoms. We have recently developed bimodal imaging framework on structural and functional network analysis and provided insight into how changes in the efficiency of structural and functional connectivity can provide a marker for diffuse deterioration in the complex connections in the brain.
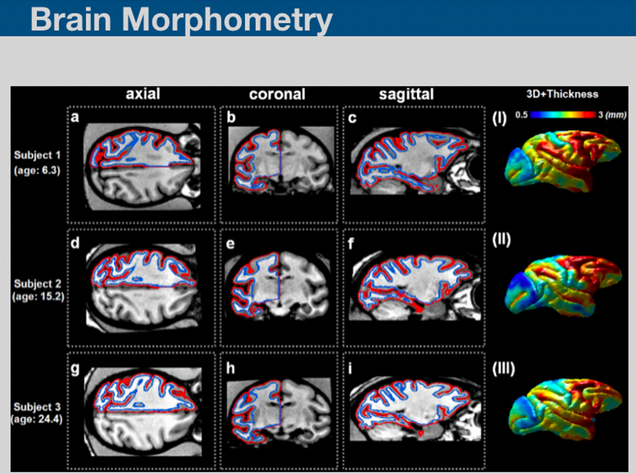
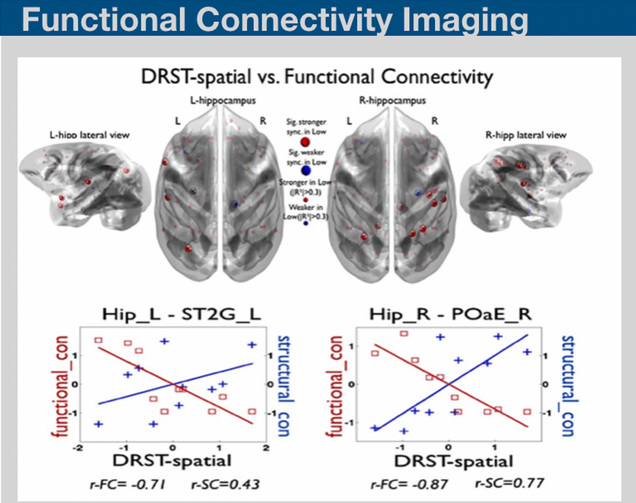
Brain morphometry is a technique involving the quantification of structural characteristics of the brain, and changes thereof, in individual or group subjects. Brain morphometry has successfully shown a detailed figure of morphological changes in the brain and its possible relationships to various psychological, behavioral, clinical or demographic factors. We are developing brain morphometry techniques fro studying animal models.
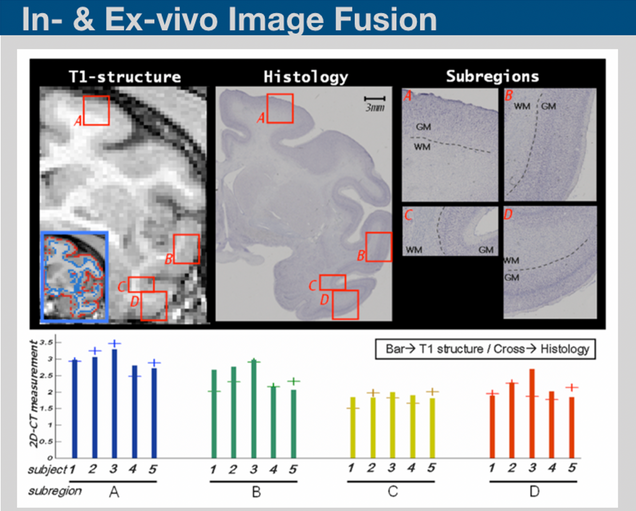
Histological sectioning and MRI are two complementary techniques that can provide both ultra micro-scale validation on tissues and sequential high throughput in-vivo monitoring of biological processes. Interfacing in-vivo scans to the histological data requires series of image processing steps including down-sampling, linear and nonlinear transformations. Based on those processing steps, information collected from in-vivo scans and be transferred into histological scans and vice versa. We are developing in-house programs to perform such image processing steps.
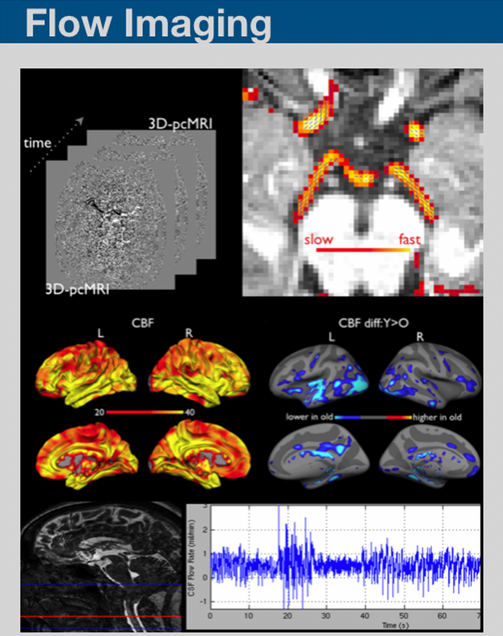
We have been working on developing MR imaging and analyses framework on imaging blood and CSF flow using pCASL, pcMRI and pencil beam imaging.
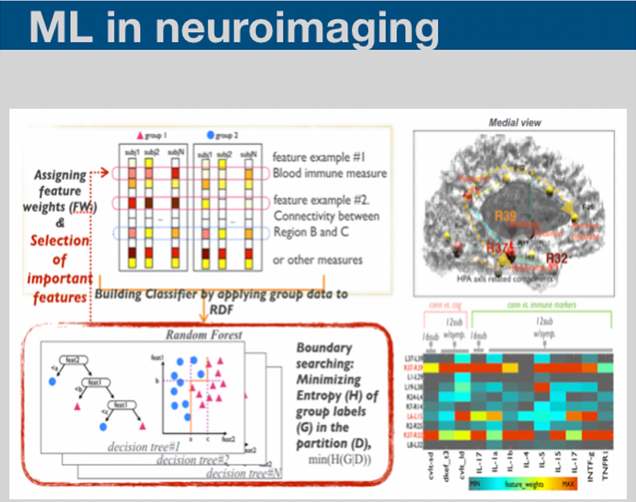
Machine learning methodologies have been essential for identifying meaningful patterns within the massive amount of data we collected. We are applying various types of machine learning methods to incorporate multiple biomarker data in order to generate the individual subject level information.