Novel Methods
Our laboratory embraces novel technologies. Below is a list of some of the novel technologies that we are using to drive our research.
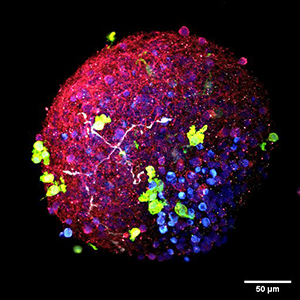
AstAD: 3D human iPSC models of AD and FTD
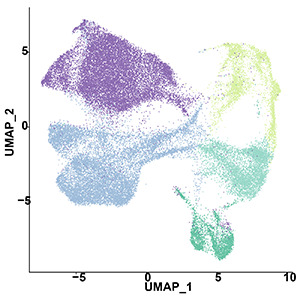
The lab has developed methods to reproduce the pathology and pathophysiology of Alzheimer’s disease using 3 dimensional assemblies of human neurons, astrocytes and microglial cells which we term AstAD. The AstAD system exhibits classic markers of tau pathology including hyper-phosphorylated tau, oligomeric tau and fibrillar tau. The AstAD system also exhibits neurodegeneration, marked by loss of MAP2 signal, increased LDH release and cleaved caspase 3. This is associated with gliosis. Single cell RNAseq studies of AstAD shows rapid, striking adaptation of ribosomal transcripts in neurons and astrocytes in response to the tau-mediated stress.
Rickner#, H.D., Jiang#, L., Hong, R., O’Neill, N. K., Wolozin*, B., Cheng*, C. S., Single cell transcriptomic profiling of neurodegeneration mediated by tau propagation in a novel 3D neuron-astrocyte coculture model. Nature Med. (2021, in review)
Optogenetics
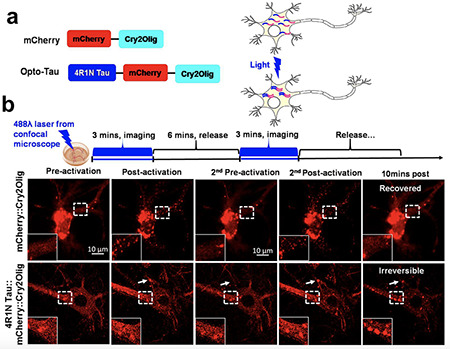
We have developed an optogenetic system for inducing tau oligomerization in a temporally controlled manner, and then combined this approach with proteomics to identify proteins that preferentially bind oTau. The optogenetics approach allows us to precisely outline the steps associated with formation of tau pathology – identifying nucleocytoplasmic translocation of TIA1 and other RNA binding proteins followed by disruption of protein synthesis and of the nuclear membrane, followed by neurodegeneration.
Lulu Jiang, Weiwei Lin, ……Andrew Emili, Benjamin Wolozin, A Complex Containing HNRNPA2B1 and N6-methyladenosine Modified Transcripts Mediates Actions of Toxic Tau Oligomers. (BIORXIV/2020/409334)
Proteomics/Emili
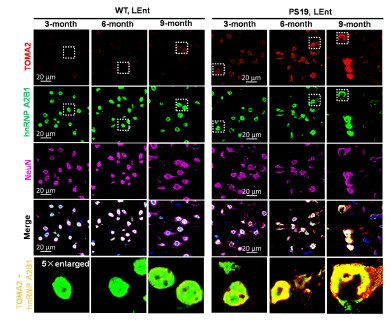
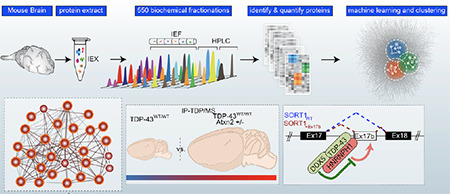
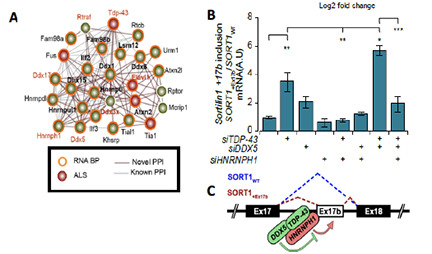
We have teamed up with the proteomics expert, Andrew Emili, to dissect the biology of AD, FTD and ALS. We have worked with the Emili group to develop BraInMap, which uses artificial intelligence and multiplexed chromatography to elucidate native protein complexes in the brain and associated with disease. This work led to the discovery that TDP-43 works in conjunction with HNRNPH1 and DDX5 to regulate splicing of key target transcripts. This work also led to discovery of the protein::protein interaction network that drives tau biology.
Lulu Jiang, Weiwei Lin, ……Andrew Emili, Benjamin Wolozin, A Complex Containing HNRNPA2B1 and N6-methyladenosine Modified Transcripts Mediates Actions of Toxic Tau Oligomers. (BIORXIV/2020/409334)
Pourhaghighi, R, Ash, PEA, ……. Wolozin, B, and Emili, A, BraInMap elucidates the macromolecular connectivity landscape of mammalian brain. Cell Syst. 2020 Apr 22;10(4):333-350.e14. doi: 10.1016/j.cels.2020.03.003. PMID: 32325033
Novel RNA species and disease
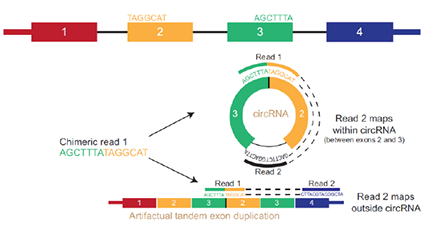
The study of RNA metabolism necessarily involves the study of many different types of RNA species. Our approaches using RNAseq and scRNAseq help to identify patterns of transcriptional change associated with disease. However, emerging studies identify novel aspects of RNA biology that appear to be strongly associated with disease. As mentioned above, are now focusing extensively on m6A to understand how it contributes to the pathophysiology of disease. We also have an emerging project examining how circular RNA contributes to the pathophysiology of neurodegenerative diseases.
Lulu Jiang, Weiwei Lin, ……Andrew Emili, Benjamin Wolozin, A Complex Containing HNRNPA2B1 and N6-methyladenosine Modified Transcripts Mediates Actions of Toxic Tau Oligomers. (BIORXIV/2020/409334)
Apicco, D. J., Zhang, C., Maziuk, B.F., Jiang, L., Balance, H.I., Boudeau, S., Ung, C., Li, H. ¶, Wolozin, B., Dysregulation of RNA splicing in tauopathies. Cell Reports 2019 Dec. 24. 29:4377-4388. PMID: 31875547. doi: 10.1016/j.celrep.2019.11.093.