Knox
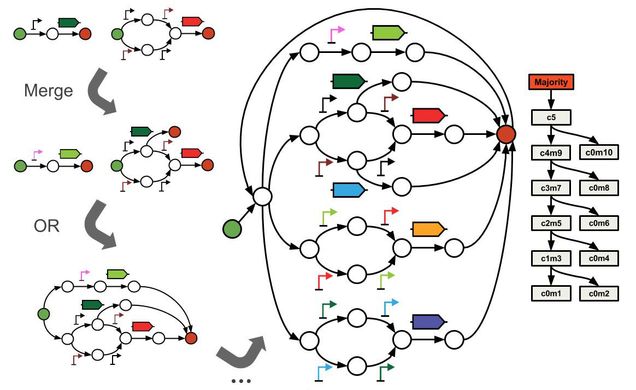
Knox is a database system for storing, making, and tracking changes to genetic design variants. Knox stores designs in a graph known as a “genetic design space.” This allows for compact storage of thousands of variants and enables new design rules to be added via graph operations such as Join, OR, and Merge. In addition, genetic design spaces can be compared via the AND operation to identify common genetic motifs. My goals for Knox are to (1) enable new software tools for learning which motifs make up successful genetic architectures and (2) to power online games that teach and learn new design rules to and from players.
Double Dutch
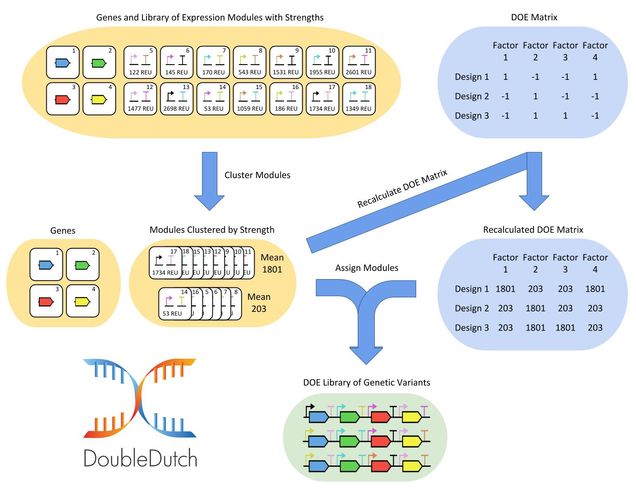
One strategy to optimize the yield of a metabolic pathway is to build and test many variants of the pathway under the control of different genetic parts. In practice, however, it can be prohibitively expensive to build a library to test all possible variants. Double Dutch is a web application that enables users to design much smaller libraries based on principles from design of experiments (DOE). These DOE libraries can be used to construct empirical models of pathway behavior and predict the set of parts that will optimize pathway yield. My plan is to incorporate Double Dutch into the design of a virtual laboratory environment that encourages users to apply DOE to their genetic designs.
Splice
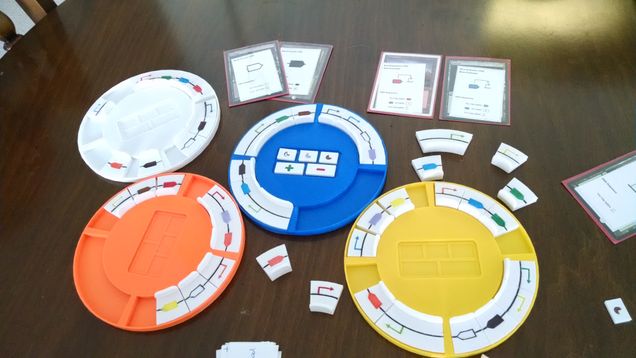

Splice is a synthetic biology board game in which players try to produce the most resources such as food, fuel, and medicine by constructing metabolic pathways and regulating their expression with genetic circuits. My goal in developing Splice was to make a game where the rules and mechanisms of play capture key concepts from synthetic biology. My playtesters and I recently demoed Splice at the 8th International Workshop on Bio-Design Automation (IWBDA) at Newcastle Upon Tyne and received positive feedback from attendees. The next step is to evaluate Splice’s potential with a slightly more general audience and make a plan for publishing the game digitally or physically. My hope is that games like Splice can aid synthetic biology outreach and give players real insight into genetic design.