New preprint: LECT2 stability and aggregation
We just posted a manuscript to the bioRxiv preprint server, “An amyloidosis-associated polymorphism does not alter LECT2 stability in vitro“. This paper describes work that was carried out in the lab by Liudmila Belonogov and Paris Taylor in 2019 and early 2020, with additional work by Sherry Wong in 2021.
There are many proteins known to aggregate as amyloid fibrils and cause human disease. One of the more recent discoveries is amyloid derived from the LECT2 protein, leukocyte chemotactic factor 2. LECT2 is a signaling protein expressed mainly in the liver, where it has roles in fibrosis and cancer, but its physiological role is not well understood. Amyloid formed from LECT2 has been found in the kidney and liver, in a disease known as amyloid LECT2 (ALECT2) amyloidosis.
The causes of ALECT2 amyloidosis are not known. Unlike inherited forms of transthyretin amyloidosis, there is no obvious mutation that causes disease. Instead, most (or possibly all) individuals are homozygous for a valine (V) residue at position 40 in the LECT2 protein sequence, which can be either valine or isoleucine (I) in the general population. This is not a rare mutation – around a sixth of people have two valine alleles – but we wondered if it might be related to the disease process. Unstable transthyretin proteins are more prone to form amyloid in vitro and in vivo, so we asked whether the V40 variant of LECT2 was less stable than the I40 variant.
We expressed both proteins in E. coli and measured their folding and aggregation using fluorescence spectroscopy. Somewhat surprisingly, we do not see a big difference between the two variants. The valine and isoleucine forms of the protein are similar in every way that we measured. There are some subtle differences in the kinetics of aggregation, but both variants form amyloid at a similar rate. We can’t say for certain that there is no difference, but there are no obvious signs that having a valine at position 40 destabilizes LECT2.
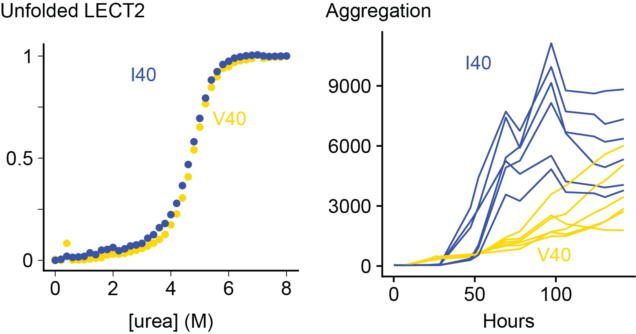
On the other hand, destabilizing LECT2 by removing a structural zinc ion made both variants aggregate much faster. So unfolding of the native state seems to be an important step towards aggregation.
By the spring of 2020, we had managed to get our initial experiments working and collected enough data to show that there are no big differences between the two proteins. However, progress was abruptly halted by the Covid-19 pandemic and we never quite managed to get the experiments restarted properly. With other priorities and funding, I made the decision to put the project on hold for the time being and make our initial data available to anyone who might be interested. It’s written into a manuscript to put the data into context, but we have no plans to submit this for peer review until we can better understand what’s going on. We’re not done with LECT2, but but I would rather have a partial story available to other researchers than let it gather dust until it’s “finished”.