A high-throughput screen for protein stability
We have developed a new method for measuring protein stability that can be used in high throughput screens. We hope that this will be a useful alternative to existing methods for researchers looking for molecules that bind to and stabilize proteins. Details are in our new paper.
Proteins are dynamic, flexible polymers that can form a range of structures. In cells, proteins are synthesized on ribosomes, acquire their functional shape (which may or may not be folded) and are eventually degraded when they are no longer needed. A new generation of drugs designed to alter the stability and lifespan of proteins is expanding the definition of a “druggable” target and making a difference in many patients’ lives. At one end of this spectrum are drugs that result in the degradation of proteins, which work by increasing the rate at which proteins are sent to the proteasome within cells. At the other end are stabilizers and pharmacological chaperones, which increase the production and lifespan of proteins that are prone to misfolding. We are working with collaborators at Scripps Research to develop molecules that stabilize antibody light chains and prevent their aggregation in AL amyloidosis.
However, rationally designing small molecules that modulate protein structure and stability, rather than enzyme activity, is difficult. Stability can mean an individual protein’s lifespan or its propensity to maintain its folded structure. These properties are intimately linked because proteins are generally removed by proteases, which preferentially degrade unfolded proteins. To measure stability in an isolated protein, we typically put it under conditions that perturb the folded state, then ask how much folded protein remains. This could be by raising the temperature, changing the pH, adding protease to digest the unfolded protein or other methods. The difficulty lies in distinguishing folded from unfolded protein. This is possible using spectroscopy or size separation, depending on how the protein was perturbed. When we measure the stability of light chains, we use fluorescence emission from the protein’s tryptophan residues or electrophoresis to separate intact from proteolysed protein.
Neither of these methods scales to the throughput needed to screen a large library of potential drug molecules – you can’t easily make the measurements on a 1536-well plate. One method that can be used is called differential scanning fluorimetry, or ThermoFluor. Here, proteins are heated until they begin to unfold. Hydrophobic dye molecules can bind to partially folded proteins, enhancing the fluorescence of the dye. As the temperature increases, dye fluorescence increases and then eventually decreases again as the protein unfolds completely. The peak of the resulting curve occurs at a protein’s thermal denaturation (“melting”) temperature. A molecule that stabilizes the protein will cause an apparent increase in the melting temperature. This can work well, but like all assays, it’s prone to interference.
We looked at using a ThermoFluor assay to find small molecules that stabilize light chains, but the initial results were not promising. So we tried another approach. Proteolysis depends on stability, so we looked for a way to ask whether a protein molecule was intact or had been cleaved by protease. We labeled light chain molecules with fluorescein, a simple fluorescent dye, and measured their fluorescence polarization as they were digested by protease.
Fluorescence polarization (also known as fluorescence anisotropy) measures rotational diffusion – the rate at which molecules tumble in solution. When a molecule absorbs light, there is a delay before that light is reemitted as fluorescence. If the molecule rotates in that time period, the polarization of the emitted light differs from that of the absorbed light. Because the molecules tumble randomly, the net polarization is averaged out if the delay between absorbance and emission is longer than the time it takes for the molecule to rotate. Small molecules tumble rapidly and larger molecules tumble more slowly, so the polarization of emitted light is averaged out more rapidly by small molecules than by large ones. This fluorescence polarization measurement can be made quickly and easily using a platereader.
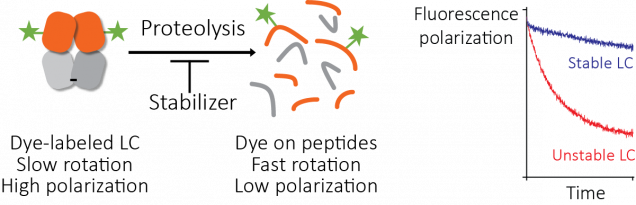
Josh Blunden, who was a summer student in the Kelly lab, was able to get this assay working. The fluorescence polarization of light chains decreases as they are degraded by protease, and the rate of degradation depends on the protein’s stability. Therefore, we could run the proteolysis reactions in parallel with different potential stabilizer molecules (650,000 in this case) to carry out our screen. We call this method the “protease-coupled fluorescence polarization”, or PCFP, assay. We found 2,777 hits that were active in the screen. Like all screens, there are ways for this to go wrong. We saw interference with the assay that seems to be from binding of dye or protein to the plate surface, but this could be solved by adding detergent or using low-binding plates. An obvious failure mode here is that the small molecule could inhibit the protease, rather than stabilize the light chain. To identify and remove any of these false positive molecules, we repeated the screen but used a different protease with a distinct catalytic mechanism (thermolysin, rather than the proteinase K that we used in the main screen). Several molecules had no effect in this assay, suggesting that they were false positive hits. One molecule was a well-characterized protease inhibitor drug, bortezomib.
After further rounds of validation and counterscreening, we are confident that the molecules described in the paper are genuine stabilizers of light chains. We are working to create more effective molecules that could become drug candidates.
We think that our PCFP method could be useful for other researchers who are looking for ways to measure stability in high throughput screens. In theory, any binding event that perturbs protease sensitivity can be measured this way. It has some advantages and disadvantages compared to a ThermoFluor assay, so it may work better in some cases. The PCFP assay is a single read in a polarization-capable platereader, whereas a ThermoFluor assay requires a long process in a qPCR instrument. PCFP may be less prone to artifacts such as protein aggregation, evaporation or dye displacement. However, it requires site-specific labeling of the protein, which can be difficult to optimize, whereas a ThermoFluor assay uses unmodified protein. The chemistry and optical readout are distinct from the ThermoFluor assay, so the two methods should be complementary. This means that true positive hit molecules should be active in both assays, while false positives may be more specific to one or other assay. One useful property is that the assay works at low protein concentration, which can be an advantage for screening. We were able to run the whole screen using 10 mg of fluorescein-labeled protein.
There’s also the potential to use this assay, or something similar, as a function-agnostic way to find protein binding molecules. Most binding assays require knowledge about the binding site, a known ligand or some kind of functional assay to work. For example, it’s easy to screen for protease inhibitors using fluorogenic substrates, or by looking for molecules that will displace a known inhibitor. Alternatively, cell-based assays that measure the activity of a signaling pathway or reporter gene have been used for all kinds of screens. One potential advantage of the PCFP assay is that you don’t need to know how the target protein works, or where its ligands bind (even its exact structure, as long as you can attach a fluorophore). The one thing you do need is data suggesting that the proteolysis of the target protein is stability-dependent, which wouldn’t necessarily be true for an intrinsically disordered region, for instance. It might be possible to get around that by using a more specific protease. We plan to try similar assays with other systems in order to see how far we can take the idea.
It’s a little surprising that no-one seems to have tried this before. Or maybe they have, but it didn’t work in their system and it was never published. I really hope that this method can solve someone’s problem and help move a project forwards. Please let me know if you want to try it and would like some advice.