Tagged: Human Connectome
The Mystery of the Human Connectome
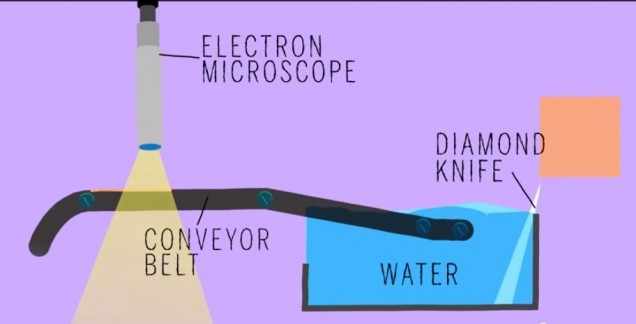
Did anyone read that article in BU today last week called “Untangling the Connectome?” In case you didn’t, here’s a brief synopsis. Dr. Kasthuri and his lab members are working on identifying the connections between the neurons in the human brain. This has been done before for C. elegans, which have only 302 neurons (compared to our 100 trillion). So, imagine how complex this project is and how much data is contained in one tiny slice of human brain?! In the article, Kasthuri said that a brain slice with the volume of “a millimeter cubed, at the resolution we would like, is about two million gigabytes of data” [1].That’s crazy. The Kasthuri lab implemented a clever method (Figure 1) to take images of 30 nm brain slices that provide the high resolution pictures they’re looking for.
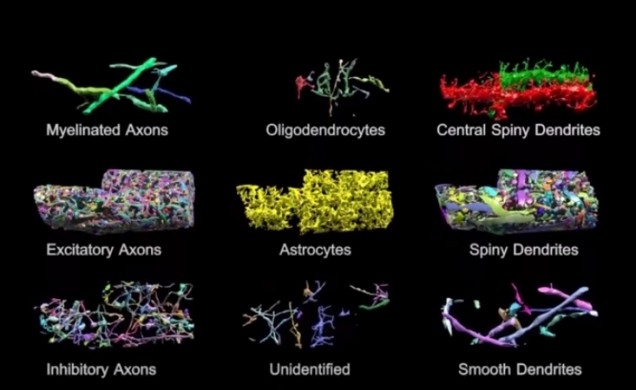
Basically, the brain volume is immersed in plastic and cut with a diamond knife to ensure precision. Since the slices are somewhat floppy, they are collected in water then picked up by a conveyor belt. While on the conveyor belt, an electron microscope takes a quick shot of the brain tissue and next a collection of images is put together to create a accurate image of the initial brain volume. Methods can be used to break down the image of the brain volume into components such as axons, dendrites, etc. (Figure 2). Pretty cool, right? The implications of this research is that understanding the connections in our brain can teach us about brain development and memory functions.
The Human Connectome project at Mass General Hospital (MGH) is working on developing high resolution neuroimaging methods. They’re working on figuring out how varying white matter fiber connectivity correlates to brain function and how cytoarchitectonics (the pattern of neuron organization/density in different areas of the brain, used to decipher Brodmann’s areas in the brain) can influence brain connectivity [2]. Methods they’re using include diffusion spectrum imaging (DSI), which is similar to diffusion tensor imaging (DTI) as both allow scientists to observe white matter connectivity since water diffuses parallel to white matter tracts. DTI measures the preferred and mean direction of the diffusion of water while DSI measures it in many different directions [3]. Scientists at MGH are also working on developing tools for high angular diffusion imaging (HARDI), which “address[es] the challenge of imaging crossing fibers by applying a stronger magnetic gradient for a longer time and capturing many more images of diffusion in various directions” [3].
Researchers are hoping to find patterns of connectivity by deciphering how our 100 trillion neurons are linked. This information could help scientists better understand the roots of different neurological or neurodegenerative diseases where perhaps neural connectivity differs significantly. This is a challenging project but the results of it could be a major breakthrough for neuroscience.
~ Srijesa K.
Sources:
[1] “Untangling the Connectome”
[3] “White’s the Matter: A basic guide to white matter imaging using diffusion MRI”