Karen Allen: Lanthanide Binding Tags
- Karen Allen
- The Allen Laboratory has pioneered and developed the use of Lanthanide binding tags (LBTs) . Since a single tag can potentially aid in protein purification, characterization, structure determination, and imaging, it may simultaneously provide new, critical tools for proteomics and medical research. LBTs are short polypeptides (15-25 amino acids) derived from calcium-binding motifs and modified to bind trivalent lanthanide ions with nM affinity. The goal is to establish stable complexes with physical properties that are useful in biochemical and biophysical investigations. Furthermore, because they are composed exclusively of amino acids, these tags can be fused to proteins of interest using standard molecular biology techniques. In the past 5 years, the Allen Laboratory has had six undergraduate researchers, including summer researchers, and their contributions have been significant. For example, the work of one student allowed the design, mutation, expression, purification and crystallization of a new tag with enhanced properties for binding the lanthanide Gd3+ and magnetic resonance imaging (MRI). The next step forward will be to: 1) design and optimize an MRI LBT fused to the cancer-related growth factor EGF in order to “light up” affected cells; and 2) to expand the repertoire of wavelengths that can be used for excitation by using “click” chemistry to attach novel sensitizes to the tags. REU students participating in this project gain experience in molecular biology as applied to protein engineering, protein purification, and X-ray crystal structure determination.
-
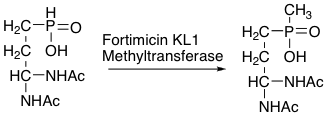
Pinghua Liu: Mechanistic Studies on C-P Bond Containing Natural Products
- Pinghua Liu
- Synthetic C-P bond containing compounds (phosphonates) have been widely used as pesticides. Phosphonates are also found in nature and most of them have biological activities. The mechanistic studies on C-P bond containing natural products in our laboratory aimed at the following two directions: 1. how the C-P bond is formed enzymatically; 2. how these phosphonates are degraded in nature. The only C-P bond formation enzyme characterized so far is the phosphoenolpyruvate mutase. The analysis of C-P containing natural product indicates that there must be other enzymatic methods to form the C-P bonds in natural products. The REU student will learn the basic molecular biology skill to clone a C-P bond formation enzyme. He/she will then work with graduate students or post doctoral scientists in the lab on organic synthesis, enzymatic activity evaluations, product purification, and enzyme inhibition studies.
-
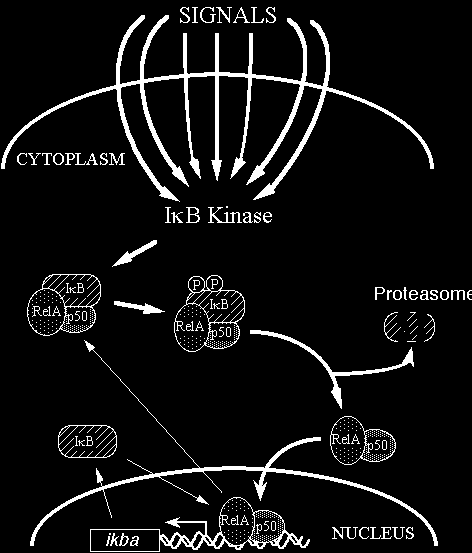
Thomas Gilmore: Structure-Function Studies of the NEMO Protein, a Key Adaptor in the NF-kB Signal Transduction Pathway
- Thomas Gilmore
- The NF-kB signal transduction pathway regulates a variety of important organismal and cellular processes, including immune responses, certain developmental processes, and cell proliferation and survival. In addition, the NF-kB pathway is misregulated in many human diseases, including a variety of inflammatory diseases and cancers. As such, there is great interest in understanding the protein components of the NF-kB pathway. One protein, NEMO, is required for NF-kB signaling in immune cells, and there are humans with immunodeficiency diseases that are caused by mutations in the NEMO protein. In collaborative studies with Professor Adrian Whitty (Chemistry), the Gilmore lab has been characterizing how human disease mutations in NEMO affect its activity and its biochemical and biophysical properties. An additional goal of these studies is to identify methods for screening for drugs that specifically target the NEMO protein, in order to modify its activity for disease therapy. In this project, an REU student will use a variety of cellular, molecular, and biochemical assays to characterize the effects of mutations in NEMO on biochemical properties and molecular activities of the NEMO protein. These approaches include tissue culture experiments, Western blotting, protein-protein interaction assays, site-directed mutagenesis, and protein expression in bacteria and human cells.
-
Deborah Perlstein: The Assembly and Function of Iron Sulfur Enzymes
- Deborah Perlstein
- The Perlstein lab studies proteins that assemble and utilize iron sulfur clusters. Iron sulfur clusters are enzyme cofactors required for a wide array of fundamental biochemical pathways including respiration, photosynthesis and nitrogen fixation. These essential enzyme cofactors cannot spontaneously assemble in enzyme active sites but instead require a dedicated biochemical pathway for their assembly. The Perlstein lab studies a collection of iron sulfur cluster assembly enzymes that are essential for the biosynthesis of extramitochondrial iron sulfur enzymes, like nuclear localized enzymes required for DNA synthesis and repair. We also study the biochemical function of iron sulfur proteins in radical-SAM enzymes. The radical SAM superfamily of enzymes use an iron sulfur cluster to reductively cleave S-adenosylmethionine (SAM) to generate a highly reactive radical intermediate which can then be utilized for chemically challenging transformations such as the functionalization of unactivated C-H bonds or carbon skeleton rearrangements. The Perlstein lab is investigating the function of a member of this family which can catalyze methylation of substrates at nonnucleophilic sites.
-
Jennifer Bhatnagar: Microbial Secretomes in the Environment
- Jennifer Bhatnagar
- Microorganisms regulate critical ecosystem functions that control the major pools and fluxes of carbon (C) between the atmosphere and the biosphere. Fungi do this by secreting extracellular enzymes and low-molecular weight peroxide-generating agents that decompose biopolymers in soil, as well as secondary metabolites that depolymerize cells walls of competitor fungal species. The fungal secretome is highly reactive in soils, where over 100 species of fungi can be present in a single cm2. However, it is not yet clear how the secretome varies across a diversity of species in Kingdom Fungi, how the microbial secretome and influences down-stream processes in the ecosystem – such as carbon and nitrogen cycling through soils or plant growth – or the impact of global changes (such as climate warming and pollution) reshape these processes. The objective of our research is to identify the molecular-level factors that regulate the secretome of soil fungi and bacteria and its effect on the biogeochemistry of ecosystems under human-induced environmental changes, using a combination of analytical and computational chemistry approaches. To do this, we collect soil and root samples from a variety of field sites around New England, then measure the molecular, biochemical, and biogeochemical activity of each species in the community using multi-omics techniques. In this project, the REU student would characterize the secretome and biogeochemical activity of bacteria and fungi during community interactions in soil experiencing different levels of urbanization intensity. These assays involve biochemical assays of extracellular enzyme activities, identification of secondary metabolites using analytical chemistry techniques, and quantification of the chemical properties of soil using biogeochemistry techniques. There is also the opportunity to learn about metabolic flux modeling to simulate microbial metabolic processes and their effect on the physiochemistry of soil particles in the environment.
-





