Publications
Perlstein lab publications on:
Publications from Boston University
24. Vasquez, S; Marquez, MD; Brignole, EJ; Vo, A; Kong, S; Park, C; Perlstein, DL; Drennan CL. Structural and biochemical investigation of a HEAT-repeat protein involved in the cytosolic iron-sulfur assembly pathway. Nature Communications Biology. 2023, 6, 1-12.
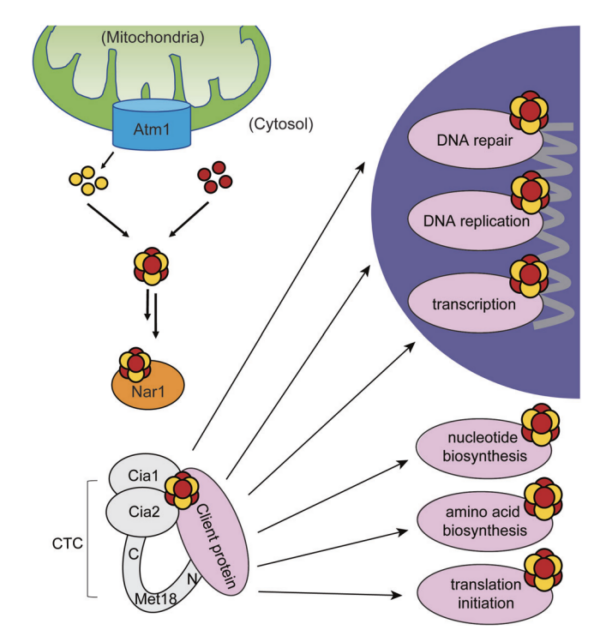
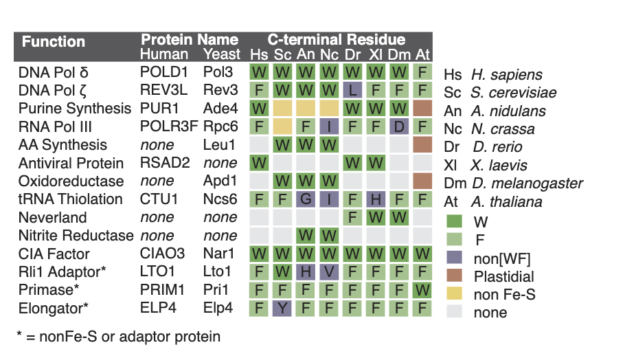
23. Marquez, MD; Greth, C; Buzuk, A; Liu, Y; Blinn, CM; Beller, S; Leiskau, L; Hushka, A; Wu, K; Nur, K; Netz, DJA; Perlstein, DL; Pierik, AJ. Cytosolic iron-sulfur protein assembly system identifies clients by a C-terminal tripeptide. PNAS. 2023, 120, 1-7.
22. Molé, CN; Dave K; Perlstein, DL. Methods to Unravel the Roles of ATPases in Fe-S Cluster Biosynthesis. Methods in Molecular Biology, 2021, 2353, 155-171.
21. Grossman, JD; Gay, KA; Camire, EJ; Walden, WE; and Perlstein, DL. Coupling of Nucleotide Binding and Hydrolysis to Iron-Sulfur Cluster Acquisition and Transfer Revealed through Genetic Dissection of the Nbp35 ATPase Site. Biochemistry, 2019, 58, 2017-2027.

20. Grossman, JD; Camire, EJ; Glynn, CA; Neil, CM; Seguinot, BO; and Perlstein, DL. The Cfd1 Subunit of the Nbp35-Cfd1 Iron Sulfur Cluster Scaffolding Complex Controls Nucleotide Binding. Biochemistry, 2019, 58, 1587-1595.

19. Grossman, JD; Camire, EJ; and Perlstein, DL. Approaches to Interrogate the Role of Nucleotide Hydrolysis by Metal Trafficking NTPases: The Nbp35-Cfd1 Iron-Sulfur Cluster Scaffold as a Case Study. Methods Enzymol. 2018, 599, 293-325.
18. Vo, AT; Fleischman, NM; Camire, EJ; Esonwune, SU; Grossman, JD; Gay, KA; Cosman, JA; and Perlstein, DL. Defining the domains of Cia2 required for its essential function in vivo and in vitro. Metallomics 2017, 9, 1645-1654.
17. Vo, AT; Fleischman, NM; Froehlich, MJ; Lee, CY; Cosman, JA; Glynn, CA; Hassan, ZO, and Perlstein, DL. Identifying the Protein Interactions of the Cytosolic Iron-Sulfur Cluster Targeting complex Essential for Its Assembly and Recognition of Apo-Targets. Biochemistry, 2018, 57, 2349-2358.

16. Camire, EJ; Grossman, JD; Thole, GJ; Fleischman, NM; and Perlstein, DL. The Yeast Nbp35-Cfd1 Cytosolic Iron-Sulfur Cluster Scaffold Is an ATPase. J. Biol. Chem. 2015, 290, 23793-23802.
Prior to Boston University:
15. Doud EH; Perlstein DL; Wolpert M; Cane DE; and Walker S. Two distinct mechanisms for TIM barrel prenyltransferases in bacteria. J. Am. Chem. Soc. 2011, 113, 1270-1273.
14. Perlstein D; Wang TS; Doud EH; Kahne D; Walker S. The Role of the substrate lipid in processive glycan polymerization by the peptidglycan glycosyltransferases. J. Am. Chem. Soc. 2010, 132, 48-49.
13. Lupoli TJ; Taniguchi T; Wang TS; Perlstein DL; Walker S; and Kahne DE. Studying a cell division amidase using defined peptidoglycan substrates. J. Am. Chem. Soc. 2009, 131, 18230-18231.
12.Ostash B, Doud EH, Lin C, Ostash I, Perlstein DL, Fuse S, Wolpert M, Kahne D, Walker S. Complete characterization of the seventeen step moenomycin biosynthetic pathway. Biochemistry, 2009, 48, 8830-41.
11. Perlstein DL, Zhang Y, Wang TS, Kahne DE, Walker S. The direction of glycan chain elongation by peptidoglycan glycosyltransferaes. J. Am. Chem. Soc., 2007, 129, 12674-12675.
10. Ortigosa AD*, Hristova D,* Perlstein DL,* Zhang Z, Huang M, Stubbe J. Determination of the in vivo stoichiometry of tyrosyl radical per beta-beta’ in Saccharomyces cerevisiae ribonucleotide reductase. Biochemistry, 2006, 45, 12282-12294.
9. Zhang Z, An X, Yang K, Perlstein DL, Hicks L, Kelleher N, Stubbe J, Huang M. Nuclear Localization of the Saccharomyces cerevisiae ribonucleotide reductase small subunit requires a karyopherin and a WD40 repeat protein. Proc. Natl. Acad. Sci. U.S.A., 2006, 103, 1422-1427.
8. Perlstein DL, Ge J, Ortigosa AD, Robblee JH, Zhang Z, Huang M, Stubbe J. The active form of the Saccharomyces cerevisiae ribonucleotide reductase small subunit is a heterodimer in vitro and in vivo. Biochemistry, 2005, 44, 15366-77.
7. Sommerhalter M, Voegtli WC, Perlstein DL, Ge J, Stubbe J, Rosenzweig AC. Structures of the yeast ribonucleotide reductase Rnr2 and Rnr4 homodimers. Biochemistry, 2004, 43, 7736-42.
6. Bennati M, Weber A, Antonic J, Perlstein DL, Robblee J, Stubbe J. Pulsed ELDOR spectroscopy measures the distance between the two tyrosyl radicals in the R2 subunit of the E. coli ribonucleotide reductase. J. Am. Chem. Soc., 2003, 125, 14988-14989.
5. Yao R, Zhang Z, An X, Bucci B, Perlstein DL, Stubbe J, Huang M. Subcellular localization of yeast ribonucleotide reductase regulated by the DNA replication and damage checkpoint pathways. Proc. Natl. Acad. Sci. U.S.A., 2003, 100, 6628-6633.
4. Voegtli WC, Ge J, Perlstein DL, Stubbe J, Rosenzweig AC. Structure of the yeast ribonucleotide reductase Y2Y4 heterodimer. Proc. Natl. Acad. Sci. U.S.A., 2001, 98, 10073-10078.
3. Ge J, Perlstein DL, Nguyen HH, Bar G, Griffin RG, Stubbe J. Why multiple small subunits (Y2 and Y4) for yeast ribonucleotide reductase? Toward understanding the role of Y4. Proc. Natl. Acad. Sci. U.S.A., 2001, 98, 10067-10072.
2. Nguyen HH, Ge J, Perlstein DL, Stubbe J. Purification of ribonucleotide reductase subunits Y1, Y2, Y3, and Y4 from yeast: Y4 plays a key role in diiron cluster assembly. Proc. Natl. Acad. Sci. U.S.A., 1999, 96, 12339-12344.
1. Rao, VJ; Perlstein, DL; Robbins, RJ; Lakshminarasimhan, PH; Kao, HM; Grey, CP; and Ramamurthy, V. Detection of low levels of Bronsted acidity in Na+Y and Na+X zeolites. Chem. Comm., 1998, 2, 268-270.
