New paper: Domain interactions in light chains
Our paper using NMR to study the dynamics of antibody light chains has just been published online. We collaborated with Enrico Rennella and Lewis Kay at the University of Toronto, who are experts in NMR spectroscopy, to ask how forces within and between the domains of a full-length light chain dimer contribute to the stability of the overall protein. We found that the dimer is more stable than its component domains, showing that there’s a strong cooperativity. This coupling of folding and dimerization helps to explain why light chain dimers are much more resistant to aggregation than their component domains. Our results suggest why AL amyloidosis is so rare — the dimeric structure of the light chain prevents the aggregation-prone regions from forming amyloid fibrils.
Light chains are synthesized as individual protein chains that fold into two domains, an N-terminal variable domain (orange in the figure) and a C-terminal constant domain (grey in the figure). These mostly pair with heavy chain proteins to form an antibody. Light chains are made in excess and the unpaired protein is secreted from cells, sometimes as homodimers that can be stabilized by a disulfide bond between the two chains.
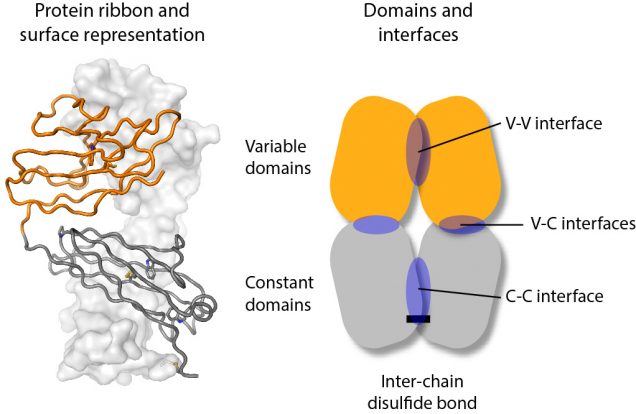
To try and understand how differences in the light chain sequence can lead to aggregation, we measured the molecular motions of a series of different light chain molecules. NMR works by measuring the resonant frequency of magnetic nuclei in an external magnetic field, which depends on the local chemistry and how that changes as molecules move in solution. We used these measurements to determine how the two protein chains of the dimer interact with each other. We looked at individual domains, full-length light chains, and a series of mutants that specifically destabilized key interactions. Although the interactions between the individual domains are individually weak, together they hold the full-length light chains together so that they form a relatively rigid molecule.
We had previously observed that full-length light chains are much less prone to aggregate than their isolated variable domains. We could now use our suite of light chain mutants to ask which interactions are necessary to prevent aggregation in native-like conditions. Light chains that could form dimers were much less likely to aggregate, but even some light chains that could only form monomers in solution remained soluble. Destabilizing the light chains’ constant domain was much more effective at causing the light chains to aggregate. This suggests that the constant domain acts to prevent aggregation of the variable domain even without the stabilizing interactions of the dimer.
We did find some surprises. First, although the two variable domains that we used have different stabilities and propensities to form dimers, there’s very little difference between how they aggregate as full-length light chains. We don’t have a good explanation for this, other than that the process of aggregation is more complex than our system is able to fully capture. Second, a mutation in the interface between the variable domains that we expected to enhance aggregation did not do so. This might be because that mutation (phenylalanine 98 to aspartic acid) also disrupts one of the regions of the light chain that forms the core of the amyloid fibril. As ever, there’s more work to be done to figure this out.
What about the bigger picture? This work supports our idea that stabilizing the full-length, natively folded light chain could help to treat AL amyloidosis. What’s encouraging is that it suggests that a small molecule that binds to any part of the light chain will stabilize the whole molecule against unfolding and aggregation. There should be more news on this story soon.