Approach
To achieve our goal, we bring together two emerging technologies:
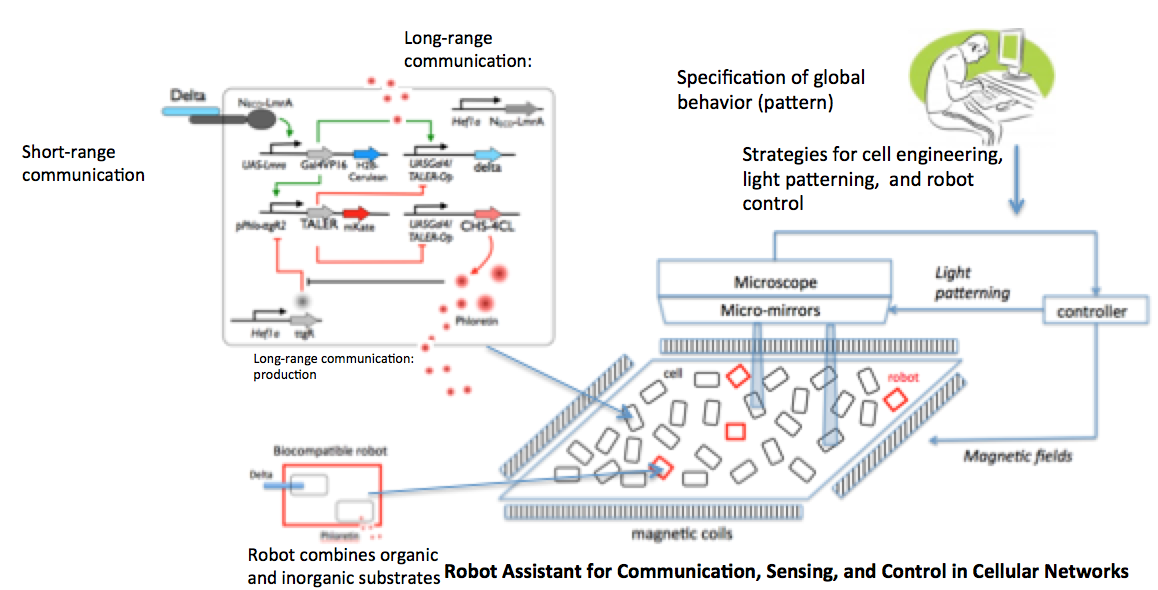
Synthetic biology: to equip the cells with sensing, communication, and decision making capabilities. Gene networks (circuits) with desired function are designed, built, and insert them into living cells.
Micron-scale robotics: to optimize the formation of patterns by affecting communication and / or moving cells and beads. Micro-scale robots are actuated using global / local bio-compatible magnetic fields, with / without magnetic levitation. Additional control is provided through opto-genetic triggering of gene circuits.
Here is a video describing our technique. More details can be found in the following sections:
Learning Patterns
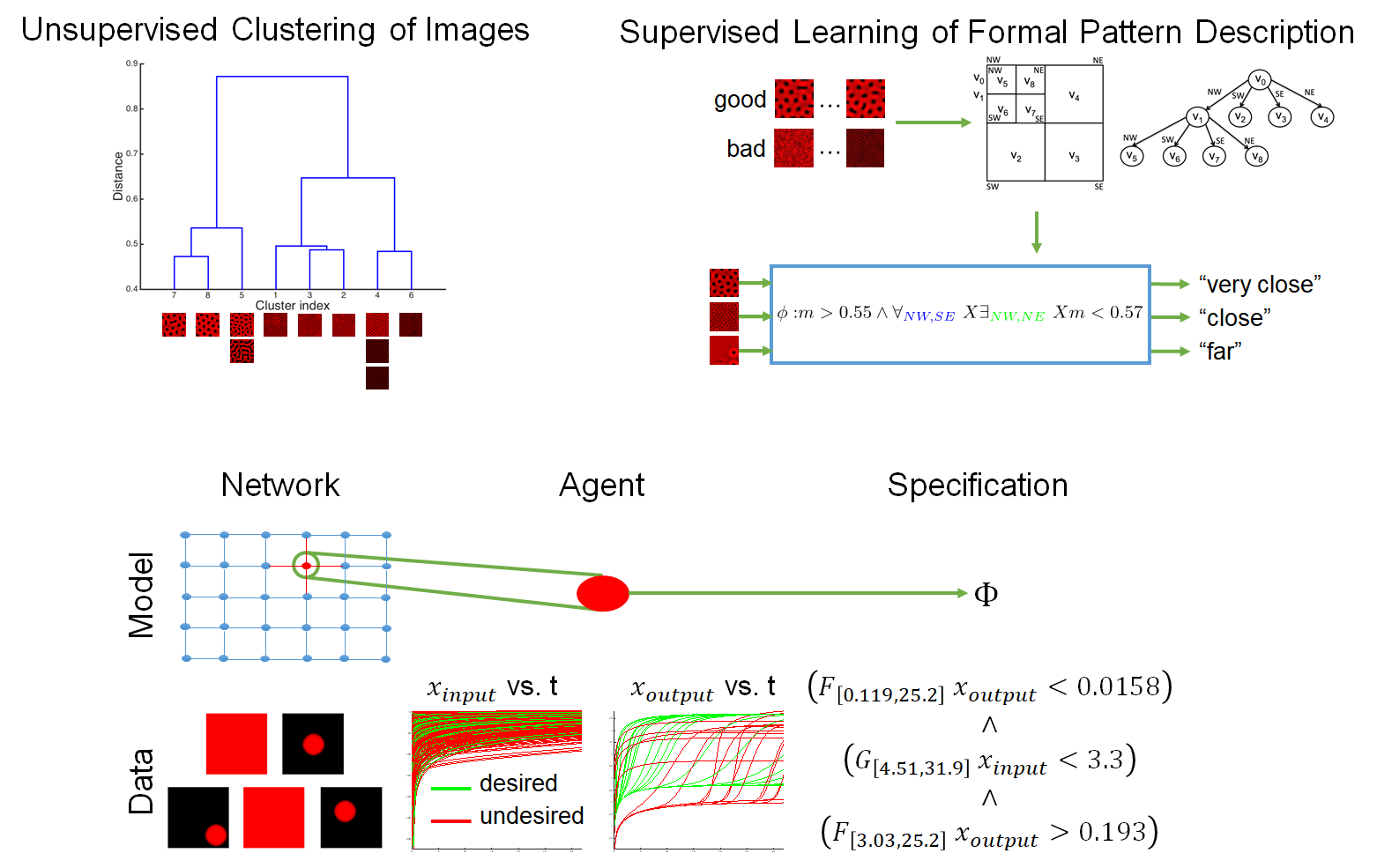
To learn a formula from a set of images, we classify the images using unsupervised learning and label them as either “desired” or “undesired”. The trajectories are labeled with respect to their image, and a heuristic supervised learning algorithm for temporal logic inference is used to build a formula in signal temporal logic. This formula attempts to describe all possible behaviors of a cell that comes from the desired pattern, i.e., if a cell helped form the desired pattern, then its trajectory will satisfy the formula. To test whether the converse is true–that all (or at least most) trajectories that satisfy the formula will help lead to the desired global pattern, we use mixed-integer linear programming to sample satisfying trajectories and examine the results. If a disproportionate number of trajectories do not lead to the desired pattern, those are added to the “undesired” group, and learning begins again. Once a formula is found that sufficiently describes the pattern, that formula is used to synthesize genetic circuits.
Bio-design Automation
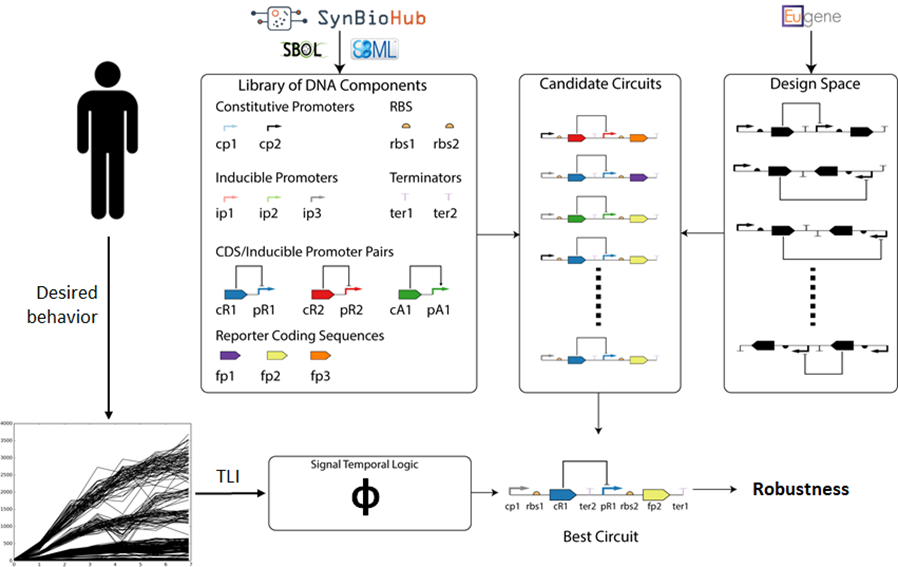
We have developed a bio-design automation (BDA) workflow that utilizes Signal Temporal Logic (STL) to specify, design, and test models of synthetic genetic circuits. With this approach, biologists can create formal, performance-bound specifications for complex genetic circuits and run finite-time simulations for modular designs to identify genetic circuits with high likelihoods of satisfying the desired specification. This workflow executes an iterative process that automatically produces circuit designs based on a specify-design-verify framework. To accomplish this goal, a user provides a set of desired behaviors as traces that are converted to STL with TLI, a structural specification, and a library of parts. The design space is searched to build candidate circuits that are verified against the STL to produce some robustness or degree of satisfaction. Finally, the circuit with the best satisfaction rate is returned.
Modeling
Biological Modules are defined by a coding sequence and a promoter. The protein that is produced by the coding sequence either activates or represses the promoter. For modules of this type, the input is the protein and/or any small molecule inducers that have an affect on this interaction, and the output is the rate of production caused by the promoter. In bacterial systems, these modules are modeled using hill-functions, and in mammalian systems, these modules are modeled using bin-dependent models. The models are constructed in such a way that they are easily composable yielding models of potential genetic circuits.
Multicellular Simulation
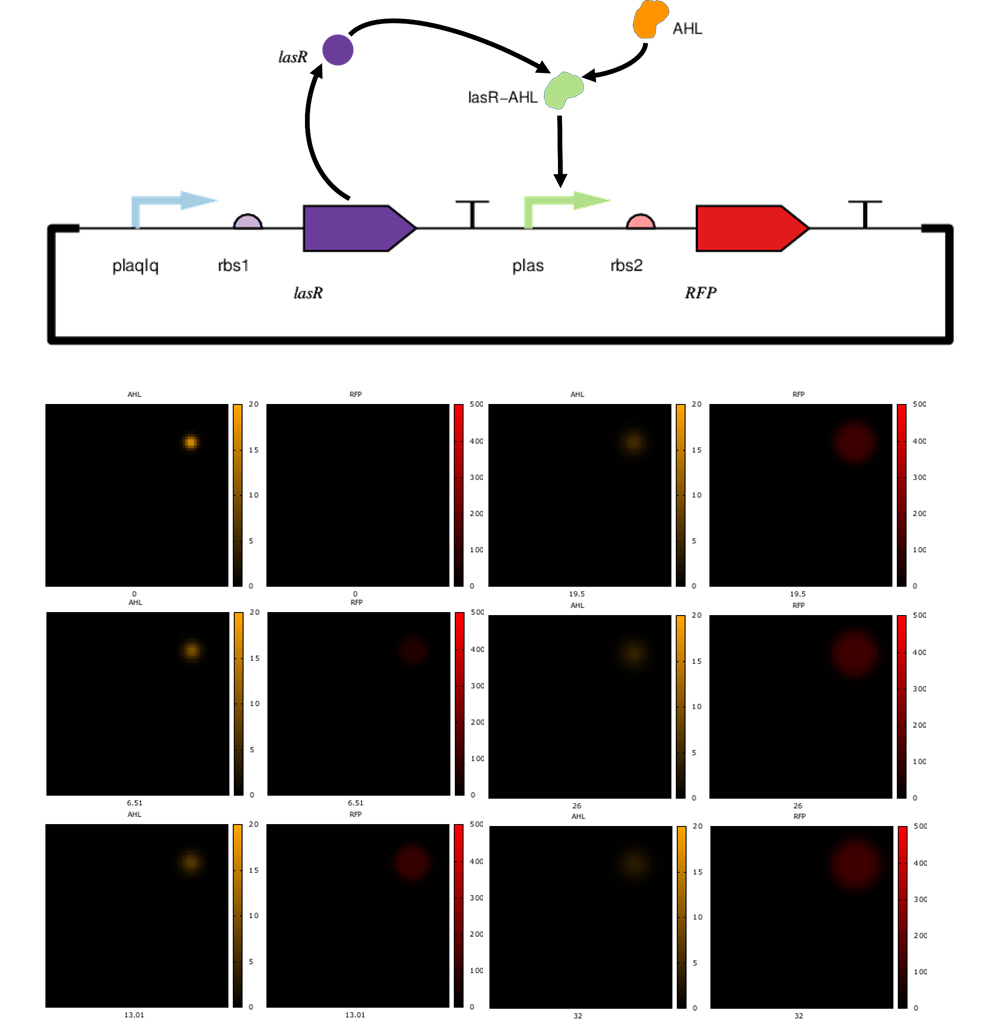
Spatial simulations of a population of cells with a circuit that turns ON in the presence of AHL producing an expanding dot. AHL is introduced at a point in space at time 0 and starts to slowly diffuse (left images). The cells at and around that point start producing RFP shortly after, and the red dot expands over time (right images).
Experimental Design
Traditional Turing reaction-diffusion patterns are formed by the combined action of a short-range activator and a long-range repressor (a). Appropriate tuning of biological and physical parameters can give rise to patterns in cell response, such as the micrographs of fluorescent E. coli in (b).
Microrobots
We explore microrobot-based cellular placement and actuation using magnetic manipulation (collaboration with SRI International). In this image, multiple microrobots are controlled in the same workplace by combining their magnetic actuation with catalytic motion. This is achieved through motion differentiation through the use of paramagnetic microrobots which assume different magnetizations in the same field. A catalytic cap helps regulate the direction of motion of the microrobot allowing for independent motion and steering control. Additionally, microrobots can be manufactured in helical structures, which are propelled via uniform rotating magnetic fields. This allows for the robot to “swim” through media spanning orders of magnitude in viscosities.